
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase, transferase
|
 |
|
Title:
|
 |
Crystal structure of the bifunctional catalytic fragment of relseq, the rela/spot homolog from streptococcus equisimilis.
|
|
Structure:
|
 |
Bifunctional rela/spot. Chain: a, b. Fragment: (p)ppgpp-3'-pyrophosphohydrolase and (p)ppgpp-synthetase subdomains. Synonym: atp:gtp 3'- pyrophosphotransferase, ppgpp synthetase i, p, ppgpp synthetase, stringent response-like protein. Engineered: yes
|
|
Source:
|
 |
Streptococcus dysgalactiae subsp. Equisimilis. Organism_taxid: 119602. Strain: subsp. Equisimilis. Gene: rela, rel, spot, rsh. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.238
|
R-free:
|
0.272
|
|
|
Authors:
|
 |
T.Hogg,U.Mechold,H.Malke,M.Cashel,R.Hilgenfeld
|
Key ref:

|
 |
T.Hogg
et al.
(2004).
Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected].
Cell,
117,
57-68.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
03-Feb-04
|
Release date:
|
04-May-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B:
E.C.2.7.6.5
- Gtp diphosphokinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
GTP + ATP = guanosine 3'-diphosphate 5'-triphosphate + AMP
|
 |
 |
 |
 |
 |
 GTP
GTP
|
+
|
ATP
Bound ligand (Het Group name = )
matches with 87.50% similarity
|
=
|
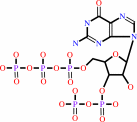 guanosine 3'-diphosphate 5'-triphosphate
guanosine 3'-diphosphate 5'-triphosphate
|
+
|
AMP
Bound ligand (Het Group name = )
matches with 77.50% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B:
E.C.3.1.7.2
- guanosine-3',5'-bis(diphosphate) 3'-diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
guanosine 3',5'-bis(diphosphate) + H2O = GDP + diphosphate + H+
|
 |
 |
 |
 |
 |
guanosine 3',5'-bis(diphosphate)
Bound ligand (Het Group name = )
matches with 86.11% similarity
|
+
|
H2O
|
=
|
 GDP
GDP
|
+
|
diphosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Cell
117:57-68
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected].
|
|
T.Hogg,
U.Mechold,
H.Malke,
M.Cashel,
R.Hilgenfeld.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Enzymes of the Rel/Spo family enable bacteria to survive prolonged periods of
nutrient limitation by producing an intracellular signaling alarmone, (p)ppGpp,
which triggers the so-called stringent response. Both the synthesis of (p)ppGpp
from ATP and GDP(GTP), and its hydrolysis to GDP(GTP) and pyrophosphate, are
catalyzed by Rel/Spo proteins. The 2.1 A crystal structure of the bifunctional
catalytic fragment of the Rel/Spo homolog from Streptococcus dysgalactiae subsp.
equisimilis, Rel(Seq), reveals two conformations of the enzyme corresponding to
known reciprocal activity states: (p)ppGpp-hydrolase-OFF/(p)ppGpp-synthetase-ON
and hydrolase-ON/synthetase-OFF. The hydrolase and synthetase domains bear
remarkable similarities to the catalytic domains of the cyclic phosphodiesterase
and nucleotidyltransferase superfamilies, respectively. The active sites,
separated by more than 30 A, contain bound nucleotides including an unusual
(p)ppGpp derivative, GDP-2':3'-cyclic monophosphate. Reciprocal regulation of
the antagonistic catalytic activities, suggested by the structure, is supported
by mutagenesis experiments and appears to involve ligand-induced signal
transmission between the two active sites.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Similarities between the Catalytic Domains of
Rel[Seq], Human Phosphodiesterase (PDE) and Human DNA Polymerase
Beta (pol β)Structural and topological diagrams highlighting
equivalent folds and active-site residues for: (A) catalytic
domain (residues 152−528) of PDE4; (B) Rel[Seq]1–385; (C)
catalytic domain (residues 10−335) of pol β. Homologous
structural elements are displayed as ribbons; nonequivalent
regions as thin gray lines. Monomer 2 of Rel[Seq]1–385 is
shown, with ppG2′:3′p, and GDP. Dark blue sphere,
catalytic metal ion (Zn^2+ for PDE4; Mn^2+ for Rel[Seq]).
Conserved residues of the H−X[(n)]−HD−X[(n)]−D metal
binding tetrad are labeled in the accompanying topology diagrams
(A and B). Two of the three catalytic carboxylates in pol β
(Asp190 and Asp256, C) are also found in Rel[Seq] (Asp264 and
Glu323, B). Rel/Spo enzymes lack a counterpart for the second
carboxylate of the D-X-D motif in NTases (Asp192 in pol β).
|
 |
Figure 4.
Figure 4. Conformations of the Synthetase Site in
Rel[Seq]1–385 and a Superposition with pol β(A) The
synthetase-ON conformation (monomer 1). Coloring is according to
Figure 1; individual structural elements are labeled in red. The
nucleophilic O3′ of GDP is marked. The final 2mFo-DFc
electron density map, shown for GDP (blue mesh), is contoured at
1.0 σ. Selected H-bonds are shown as gray dashed lines. The
catalytic loop (α13/β4) harboring Asp264 is stabilized in a
3[10]-helical conformation through multiple van der Waals
interactions (represented by black double-arrow dashed lines)
with loop α11/α12 and the first two turns of α12 (labeled t1,
t2). Chain traces extending from loops α11/α12, β3/α13, and
α13/β4 are not visible due to image slab restrictions.(B) The
synthetase-OFF conformation (monomer 2). The α11/α12 loop and
the first two turns of α12 are disordered; the resulting
elimination of van der Waals contacts to the catalytic loop
(α13/β4) leads to (1), partial refolding of the latter into an
N-terminal extension of β4, and (2), disorder of residues
254–261 including the remaining residues of the catalytic loop
and the C terminus of α13.(C) Representative electron density
in the synthetase site of monomer 1. GDP is highlighted in
orange. The final 2mFo-DFc electron density map (1.0 σ) is
overlaid as blue mesh.(D) Stereographic superposition between
the synthetase site of Rel[Seq]1–385 (monomer 1) and the
active site of pol β in the (pol β)·(gapped
DNA)·(ddCTP) complex. The latter complex is rendered in
gray shading with the exception of the primer 3′-terminal
nucleotide (orange), ddCTP (cyan), and the two Mg^2+ ions (dark
blue). Rel[Seq]1–385 and its GDP ligand are colored according
to (A). The putative catalytic carboxylates of Rel[Seq], Asp264
and Glu323, are N terminally frameshifted by two residues
relative to their pol β counterparts (indicated by black
arrows).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Cell
(2004,
117,
57-68)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.C.Boutte,
and
S.Crosson
(2011).
The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment.
|
| |
Mol Microbiol,
80,
695-714.
|
 |
|
|
|
|
 |
D.Ning,
Y.Qian,
X.Miao,
and
C.Wen
(2011).
Role of the all1549 (ana-rsh) Gene, A relA/spoT Homolog, of the Cyanobacterium Anabaena sp. PCC7120.
|
| |
Curr Microbiol,
62,
1767-1773.
|
 |
|
|
|
|
 |
P.Stevens,
L.S.van Overbeek,
and
J.D.van Elsas
(2011).
Ralstonia solanacearum ΔPGI-1 strain KZR-5 is affected in growth, response to cold stress and invasion of tomato.
|
| |
Microb Ecol,
61,
101-112.
|
 |
|
|
|
|
 |
D.Sun,
G.Lee,
J.H.Lee,
H.Y.Kim,
H.W.Rhee,
S.Y.Park,
K.J.Kim,
Y.Kim,
B.Y.Kim,
J.I.Hong,
C.Park,
H.E.Choy,
J.H.Kim,
Y.H.Jeon,
and
J.Chung
(2010).
A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses.
|
| |
Nat Struct Mol Biol,
17,
1188-1194.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Geiger,
C.Goerke,
M.Fritz,
T.Schäfer,
K.Ohlsen,
M.Liebeke,
M.Lalk,
and
C.Wolz
(2010).
Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus.
|
| |
Infect Immun,
78,
1873-1883.
|
 |
|
|
|
|
 |
W.Gao,
K.Chua,
J.K.Davies,
H.J.Newton,
T.Seemann,
P.F.Harrison,
N.E.Holmes,
H.W.Rhee,
J.I.Hong,
E.L.Hartland,
T.P.Stinear,
and
B.P.Howden
(2010).
Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection.
|
| |
PLoS Pathog,
6,
e1000944.
|
 |
|
|
|
|
 |
Y.Zhang,
E.L.Pohlmann,
J.Serate,
M.C.Conrad,
and
G.P.Roberts
(2010).
Mutagenesis and functional characterization of the four domains of GlnD, a bifunctional nitrogen sensor protein.
|
| |
J Bacteriol,
192,
2711-2721.
|
 |
|
|
|
|
 |
A.Battesti,
and
E.Bouveret
(2009).
Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction.
|
| |
J Bacteriol,
191,
616-624.
|
 |
|
|
|
|
 |
A.Boehm,
S.Steiner,
F.Zaehringer,
A.Casanova,
F.Hamburger,
D.Ritz,
W.Keck,
M.Ackermann,
T.Schirmer,
and
U.Jenal
(2009).
Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress.
|
| |
Mol Microbiol,
72,
1500-1516.
|
 |
|
|
|
|
 |
A.Danchin
(2009).
Bacteria as computers making computers.
|
| |
FEMS Microbiol Rev,
33,
3.
|
 |
|
|
|
|
 |
B.Das,
R.R.Pal,
S.Bag,
and
R.K.Bhadra
(2009).
Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p)ppGpp synthetase gene.
|
| |
Mol Microbiol,
72,
380-398.
|
 |
|
|
|
|
 |
H.M.Gan,
L.Buckley,
E.Szegedi,
A.O.Hudson,
and
M.A.Savka
(2009).
Identification of an rsh gene from a Novosphingobium sp. necessary for quorum-sensing signal accumulation.
|
| |
J Bacteriol,
191,
2551-2560.
|
 |
|
|
|
|
 |
K.M.Kazmierczak,
K.J.Wayne,
A.Rechtsteiner,
and
M.E.Winkler
(2009).
Roles of rel(Spn) in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39.
|
| |
Mol Microbiol,
72,
590-611.
|
 |
|
|
|
|
 |
K.Podzelinska,
S.M.He,
M.Wathier,
A.Yakunin,
M.Proudfoot,
B.Hove-Jensen,
D.L.Zechel,
and
Z.Jia
(2009).
Structure of PhnP, a Phosphodiesterase of the Carbon-Phosphorus Lyase Pathway for Phosphonate Degradation.
|
| |
J Biol Chem,
284,
17216-17226.
|
 |
|
|
|
|
 |
M.Sajish,
S.Kalayil,
S.K.Verma,
V.K.Nandicoori,
and
B.Prakash
(2009).
The significance of EXDD and RXKD motif conservation in Rel proteins.
|
| |
J Biol Chem,
284,
9115-9123.
|
 |
|
|
|
|
 |
R.Mega,
N.Kondo,
N.Nakagawa,
S.Kuramitsu,
and
R.Masui
(2009).
Two dNTP triphosphohydrolases from Pseudomonas aeruginosa possess diverse substrate specificities.
|
| |
FEBS J,
276,
3211-3221.
|
 |
|
|
|
|
 |
T.Ooga,
Y.Ohashi,
S.Kuramitsu,
Y.Koyama,
M.Tomita,
T.Soga,
and
R.Masui
(2009).
Degradation of ppGpp by nudix pyrophosphatase modulates the transition of growth phase in the bacterium thermus thermophilus.
|
| |
J Biol Chem,
284,
15549-15556.
|
 |
|
|
|
|
 |
T.Schirmer,
and
U.Jenal
(2009).
Structural and mechanistic determinants of c-di-GMP signalling.
|
| |
Nat Rev Microbiol,
7,
724-735.
|
 |
|
|
|
|
 |
A.Srivatsan,
and
J.D.Wang
(2008).
Control of bacterial transcription, translation and replication by (p)ppGpp.
|
| |
Curr Opin Microbiol,
11,
100-105.
|
 |
|
|
|
|
 |
B.Das,
and
R.K.Bhadra
(2008).
Molecular characterization of vibrio cholerae DeltarelA DeltaspoT double mutants.
|
| |
Arch Microbiol,
189,
227-238.
|
 |
|
|
|
|
 |
B.Spira,
X.Hu,
and
T.Ferenci
(2008).
Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12.
|
| |
Microbiology,
154,
2887-2895.
|
 |
|
|
|
|
 |
J.A.Lesley,
and
L.Shapiro
(2008).
SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus.
|
| |
J Bacteriol,
190,
6867-6880.
|
 |
|
|
|
|
 |
K.Potrykus,
and
M.Cashel
(2008).
(p)ppGpp: still magical?
|
| |
Annu Rev Microbiol,
62,
35-51.
|
 |
|
|
|
|
 |
M.D.Zimmerman,
M.Proudfoot,
A.Yakunin,
and
W.Minor
(2008).
Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5'-deoxyribonucleotidase YfbR from Escherichia coli.
|
| |
J Mol Biol,
378,
215-226.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.M.Nascimento,
J.A.Lemos,
J.Abranches,
V.K.Lin,
and
R.A.Burne
(2008).
Role of RelA of Streptococcus mutans in global control of gene expression.
|
| |
J Bacteriol,
190,
28-36.
|
 |
|
|
|
|
 |
N.Kondo,
T.Nishikubo,
T.Wakamatsu,
H.Ishikawa,
N.Nakagawa,
S.Kuramitsu,
and
R.Masui
(2008).
Insights into different dependence of dNTP triphosphohydrolase on metal ion species from intracellular ion concentrations in Thermus thermophilus.
|
| |
Extremophiles,
12,
217-223.
|
 |
|
|
|
|
 |
R.Harinarayanan,
H.Murphy,
and
M.Cashel
(2008).
Synthetic growth phenotypes of Escherichia coli lacking ppGpp and transketolase A (tktA) are due to ppGpp-mediated transcriptional regulation of tktB.
|
| |
Mol Microbiol,
69,
882-894.
|
 |
|
|
|
|
 |
S.Lee,
M.H.Kim,
B.S.Kang,
J.S.Kim,
G.H.Kim,
Y.G.Kim,
and
K.J.Kim
(2008).
Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
|
| |
J Biol Chem,
283,
15232-15240.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Bougdour,
and
S.Gottesman
(2007).
ppGpp regulation of RpoS degradation via anti-adaptor protein IraP.
|
| |
Proc Natl Acad Sci U S A,
104,
12896-12901.
|
 |
|
|
|
|
 |
J.A.Lemos,
V.K.Lin,
M.M.Nascimento,
J.Abranches,
and
R.A.Burne
(2007).
Three gene products govern (p)ppGpp production by Streptococcus mutans.
|
| |
Mol Microbiol,
65,
1568-1581.
|
 |
|
|
|
|
 |
M.Jiang,
S.M.Sullivan,
P.K.Wout,
and
J.R.Maddock
(2007).
G-protein control of the ribosome-associated stress response protein SpoT.
|
| |
J Bacteriol,
189,
6140-6147.
|
 |
|
|
|
|
 |
M.Sajish,
D.Tiwari,
D.Rananaware,
V.K.Nandicoori,
and
B.Prakash
(2007).
A charge reversal differentiates (p)ppGpp synthesis by monofunctional and bifunctional Rel proteins.
|
| |
J Biol Chem,
282,
34977-34983.
|
 |
|
|
|
|
 |
N.Kondo,
N.Nakagawa,
A.Ebihara,
L.Chen,
Z.J.Liu,
B.C.Wang,
S.Yokoyama,
S.Kuramitsu,
and
R.Masui
(2007).
Structure of dNTP-inducible dNTP triphosphohydrolase: insight into broad specificity for dNTPs and triphosphohydrolase-type hydrolysis.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
230-239.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Battesti,
and
E.Bouveret
(2006).
Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism.
|
| |
Mol Microbiol,
62,
1048-1063.
|
 |
|
|
|
|
 |
D.H.Wells,
and
E.C.Gaynor
(2006).
Helicobacter pylori initiates the stringent response upon nutrient and pH downshift.
|
| |
J Bacteriol,
188,
3726-3729.
|
 |
|
|
|
|
 |
J.Alvarado,
A.Ghosh,
T.Janovitz,
A.Jauregui,
M.S.Hasson,
and
D.A.Sanders
(2006).
Origin of exopolyphosphatase processivity: Fusion of an ASKHA phosphotransferase and a cyclic nucleotide phosphodiesterase homolog.
|
| |
Structure,
14,
1263-1272.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Braeken,
M.Moris,
R.Daniels,
J.Vanderleyden,
and
J.Michiels
(2006).
New horizons for (p)ppGpp in bacterial and plant physiology.
|
| |
Trends Microbiol,
14,
45-54.
|
 |
|
|
|
|
 |
K.Kasai,
T.Nishizawa,
K.Takahashi,
T.Hosaka,
H.Aoki,
and
K.Ochi
(2006).
Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus.
|
| |
J Bacteriol,
188,
7111-7122.
|
 |
|
|
|
|
 |
L.N.DiDonato,
S.A.Sullivan,
B.A.Methé,
K.P.Nevin,
R.England,
and
D.R.Lovley
(2006).
Role of RelGsu in stress response and Fe(III) reduction in Geobacter sulfurreducens.
|
| |
J Bacteriol,
188,
8469-8478.
|
 |
|
|
|
|
 |
M.Dozot,
R.A.Boigegrain,
R.M.Delrue,
R.Hallez,
S.Ouahrani-Bettache,
I.Danese,
J.J.Letesson,
X.De Bolle,
and
S.Köhler
(2006).
The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB.
|
| |
Cell Microbiol,
8,
1791-1802.
|
 |
|
|
|
|
 |
U.Jenal,
and
J.Malone
(2006).
Mechanisms of cyclic-di-GMP signaling in bacteria.
|
| |
Annu Rev Genet,
40,
385-407.
|
 |
|
|
|
|
 |
V.Jain,
R.Saleem-Batcha,
A.China,
and
D.Chatterji
(2006).
Molecular dissection of the mycobacterial stringent response protein Rel.
|
| |
Protein Sci,
15,
1449-1464.
|
 |
|
|
|
|
 |
A.Calderón-Flores,
G.Du Pont,
A.Huerta-Saquero,
H.Merchant-Larios,
L.Servín-González,
and
S.Durán
(2005).
The stringent response is required for amino acid and nitrate utilization, nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli.
|
| |
J Bacteriol,
187,
5075-5083.
|
 |
|
|
|
|
 |
D.Vinella,
C.Albrecht,
M.Cashel,
and
R.D'Ari
(2005).
Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli.
|
| |
Mol Microbiol,
56,
958-970.
|
 |
|
|
|
|
 |
H.Ogata,
P.Renesto,
S.Audic,
C.Robert,
G.Blanc,
P.E.Fournier,
H.Parinello,
J.M.Claverie,
and
D.Raoult
(2005).
The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite.
|
| |
PLoS Biol,
3,
e248.
|
 |
|
|
|
|
 |
J.D.Gralla
(2005).
Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein.
|
| |
Mol Microbiol,
55,
973-977.
|
 |
|
|
|
|
 |
L.U.Magnusson,
A.Farewell,
and
T.Nyström
(2005).
ppGpp: a global regulator in Escherichia coli.
|
| |
Trends Microbiol,
13,
236-242.
|
 |
|
|
|
|
 |
A.F.Yakunin,
M.Proudfoot,
E.Kuznetsova,
A.Savchenko,
G.Brown,
C.H.Arrowsmith,
and
A.M.Edwards
(2004).
The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2',3'-cyclic phosphodiesterase, 2'-nucleotidase, and phosphatase activities.
|
| |
J Biol Chem,
279,
36819-36827.
|
 |
|
|
|
|
 |
B.J.Paul,
M.M.Barker,
W.Ross,
D.A.Schneider,
C.Webb,
J.W.Foster,
and
R.L.Gourse
(2004).
DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP.
|
| |
Cell,
118,
311-322.
|
 |
|
|
|
|
 |
B.J.Paul,
W.Ross,
T.Gaal,
and
R.L.Gourse
(2004).
rRNA transcription in Escherichia coli.
|
| |
Annu Rev Genet,
38,
749-770.
|
 |
|
|
|
|
 |
M.Proudfoot,
E.Kuznetsova,
G.Brown,
N.N.Rao,
M.Kitagawa,
H.Mori,
A.Savchenko,
and
A.F.Yakunin
(2004).
General enzymatic screens identify three new nucleotidases in Escherichia coli. Biochemical characterization of SurE, YfbR, and YjjG.
|
| |
J Biol Chem,
279,
54687-54694.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

