 |
PDBsum entry 1v4g

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of gamma-glutamylcysteine synthetase from escherichia coli b
|
|
Structure:
|
 |
Glutamate--cysteine ligase. Chain: a, b, c, d. Synonym: gamma-glutamylcysteine synthetase, gamma-ecs, gcs. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Gene: gsh-i. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.207
|
R-free:
|
0.236
|
|
|
Authors:
|
 |
T.Hibi,H.Nii,T.Nakatsu,H.Kato,J.Hiratake,J.Oda
|
Key ref:

|
 |
T.Hibi
et al.
(2004).
Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis.
Proc Natl Acad Sci U S A,
101,
15052-15057.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
13-Nov-03
|
Release date:
|
05-Oct-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0A6W9
(GSH1_ECOLI) -
Glutamate--cysteine ligase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
518 a.a.
509 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.2.2
- glutamate--cysteine ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-cysteine + L-glutamate + ATP = gamma-L-glutamyl-L-cysteine + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
 L-cysteine
L-cysteine
|
+
|
 L-glutamate
L-glutamate
|
+
|
 ATP
ATP
|
=
|
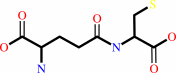 gamma-L-glutamyl-L-cysteine
gamma-L-glutamyl-L-cysteine
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
101:15052-15057
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis.
|
|
T.Hibi,
H.Nii,
T.Nakatsu,
A.Kimura,
H.Kato,
J.Hiratake,
J.Oda.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Gamma-glutamylcysteine synthetase (gammaGCS), a rate-limiting enzyme in
glutathione biosynthesis, plays a central role in glutathione homeostasis and is
a target for development of potential therapeutic agents against parasites and
cancer. We have determined the crystal structures of Escherichia coli gammaGCS
unliganded and complexed with a sulfoximine-based transition-state analog
inhibitor at resolutions of 2.5 and 2.1 A, respectively. In the crystal
structure of the complex, the bound inhibitor is phosphorylated at the
sulfoximido nitrogen and is coordinated to three Mg2+ ions. The cysteine-binding
site was identified; it is formed inductively at the transition state. In the
unliganded structure, an open space exists around the representative
cysteine-binding site and is probably responsible for the competitive binding of
glutathione. Upon inhibitor binding, the side chains of Tyr-241 and Tyr-300
turn, forming a hydrogen-bonding triad with the carboxyl group of the
inhibitor's cysteine moiety, allowing this moiety to fit tightly into the
cysteine-binding site with concomitant accommodation of its side chain into a
shallow pocket. This movement is caused by a conformational change of a switch
loop (residues 240-249). Based on this crystal structure, the cysteine-binding
sites of mammalian and parasitic gammaGCSs were predicted by multiple sequence
alignment, although no significant sequence identity exists between the E. coli
gammaGCS and its eukaryotic homologues. The identification of this
cysteine-binding site provides important information for the rational design of
novel gammaGCS inhibitors.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Stereoview of the residues surrounding the
Cys-analog moiety of sulfoximine 2, showing the distances
between the ligands. The molecular surface around the
Cys-binding site is drawn in white.
|
 |
Figure 5.
Fig. 5. Superimposition of residues 238-251, including the
switch loop. The loop's hinge residues, Gly-240 and Leu-249, are
labeled.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
F.A.Cerda-Maira,
M.J.Pearce,
M.Fuortes,
W.R.Bishai,
S.R.Hubbard,
and
K.H.Darwin
(2010).
Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis.
|
| |
Mol Microbiol,
77,
1123-1135.
|
 |
|
|
|
|
 |
B.Geissler,
A.Bonebrake,
K.L.Sheahan,
M.E.Walker,
and
K.J.Satchell
(2009).
Genetic determination of essential residues of the Vibrio cholerae actin cross-linking domain reveals functional similarity with glutamine synthetases.
|
| |
Mol Microbiol,
73,
858-868.
|
 |
|
|
|
|
 |
C.C.Franklin,
D.S.Backos,
I.Mohar,
C.C.White,
H.J.Forman,
and
T.J.Kavanagh
(2009).
Structure, function, and post-translational regulation of the catalytic and modifier subunits of glutamate cysteine ligase.
|
| |
Mol Aspects Med,
30,
86-98.
|
 |
|
|
|
|
 |
E.I.Biterova,
and
J.J.Barycki
(2009).
Mechanistic details of glutathione biosynthesis revealed by crystal structures of Saccharomyces cerevisiae glutamate cysteine ligase.
|
| |
J Biol Chem,
284,
32700-32708.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Striebel,
F.Imkamp,
M.Sutter,
M.Steiner,
A.Mamedov,
and
E.Weber-Ban
(2009).
Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes.
|
| |
Nat Struct Mol Biol,
16,
647-651.
|
 |
|
|
|
|
 |
G.T.Wondrak
(2009).
Redox-directed cancer therapeutics: molecular mechanisms and opportunities.
|
| |
Antioxid Redox Signal,
11,
3013-3069.
|
 |
|
|
|
|
 |
K.J.Satchell
(2009).
Actin Crosslinking Toxins of Gram-Negative Bacteria.
|
| |
Toxins (Basel),
1,
123-133.
|
 |
|
|
|
|
 |
K.Miyake,
and
S.Kakita
(2009).
A novel catalytic ability of gamma-glutamylcysteine synthetase of Escherichia coli and its application in theanine production.
|
| |
Biosci Biotechnol Biochem,
73,
2677-2683.
|
 |
|
|
|
|
 |
K.Yamanaka,
C.Maruyama,
H.Takagi,
and
Y.Hamano
(2008).
Epsilon-poly-L-lysine dispersity is controlled by a highly unusual nonribosomal peptide synthetase.
|
| |
Nat Chem Biol,
4,
766-772.
|
 |
|
|
|
|
 |
T.Rausch,
R.Gromes,
V.Liedschulte,
I.Müller,
J.Bogs,
V.Galovic,
and
A.Wachter
(2007).
Novel insight into the regulation of GSH biosynthesis in higher plants.
|
| |
Plant Biol (Stuttg),
9,
565-572.
|
 |
|
|
|
|
 |
D.Toroser,
C.S.Yarian,
W.C.Orr,
and
R.S.Sohal
(2006).
Mechanisms of gamma-glutamylcysteine ligase regulation.
|
| |
Biochim Biophys Acta,
1760,
233-244.
|
 |
|
|
|
|
 |
L.Masip,
K.Veeravalli,
and
G.Georgiou
(2006).
The many faces of glutathione in bacteria.
|
| |
Antioxid Redox Signal,
8,
753-762.
|
 |
|
|
|
|
 |
J.Hiratake
(2005).
Enzyme inhibitors as chemical tools to study enzyme catalysis: rational design, synthesis, and applications.
|
| |
Chem Rec,
5,
209-228.
|
 |
|
|
|
|
 |
P.M.Mullineaux,
and
T.Rausch
(2005).
Glutathione, photosynthesis and the redox regulation of stress-responsive gene expression.
|
| |
Photosynth Res,
86,
459-474.
|
 |
|
|
|
|
 |
W.W.Krajewski,
T.A.Jones,
and
S.L.Mowbray
(2005).
Structure of Mycobacterium tuberculosis glutamine synthetase in complex with a transition-state mimic provides functional insights.
|
| |
Proc Natl Acad Sci U S A,
102,
10499-10504.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

