 |
PDBsum entry 1v48

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.1
- purine-nucleoside phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha- D-ribose 1-phosphate
|
|
2.
|
a purine 2'-deoxy-D-ribonucleoside + phosphate = a purine nucleobase + 2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
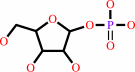 alpha- D-ribose 1-phosphate
alpha- D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine 2'-deoxy-D-ribonucleoside
|
+
|
 phosphate
phosphate
|
=
|
purine nucleobase
|
+
|
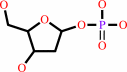 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
60:1417-1424
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Calf spleen purine-nucleoside phosphorylase: crystal structure of the binary complex with a potent multisubstrate analogue inhibitor.
|
|
M.Luić,
G.Koellner,
T.Yokomatsu,
S.Shibuya,
A.Bzowska.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Purine-nucleoside phosphorylase (PNP) deficiency in humans leads to inhibition
of the T-cell response. Potent membrane-permeable inhibitors of this enzyme are
therefore considered to be potential immunosuppressive agents. The binary
complex of the trimeric calf spleen phosphorylase, which is highly homologous to
human PNP, with the potent ground-state analogue inhibitor
9-(5,5-difluoro-5-phosphonopentyl)guanine (DFPP-G) was crystallized in the cubic
space group P2(1)3, with unit-cell parameter a = 93.183 A and one monomer per
asymmetric unit. High-resolution X-ray diffraction data were collected using
synchrotron radiation (EMBL Outstation, DESY, Hamburg, station X13). The crystal
structure was refined to a resolution of 2.2 A and R and Rfree values of 19.1
and 24.2%, respectively. The crystal structure confirms that DFPP-G acts as a
multisubstrate analogue inhibitor as it binds to both nucleoside- and
phosphate-binding sites. The structure also provides the answers to some
questions regarding the substrate specificity and molecular mechanism of
trimeric PNPs. The wide access to the active-site pocket that was observed in
the reported structure as a result of the flexibility or disorder of two loops
(residues 60-65 and 251-266) strongly supports the random binding of substrates.
The putative hydrogen bonds identified in the base-binding site indicate that
N1-H and not O6 of the purine base defines the specificity of trimeric PNPs.
This is confirmed by the fact that the contact of guanine O6 with Asn243 Odelta1
is not a direct contact but is mediated by a water molecule. Participation of
Arg84 in the binding of the phosphonate group experimentally verifies the
previous suggestion [Blackburn & Kent (1986), J. Chem. Soc. Perkin Trans. I,
pp. 913-917; Halazy et al. (1991), J. Am. Chem. Soc. 113, 315-317] that
fluorination of alkylphosphonates yields compounds with properties that suitably
resemble those of phosphate esters and in turn leads to optimized interactions
of such analogues with the phosphate-binding site residues. DFPP-G shows a
Ki(app) in the nanomolar range towards calf and human PNPs. To date, no
high-resolution X-ray structures of these enzymes with such potent ground-state
analogue inhibitors have been available in the Protein Data Bank. The present
structure may thus be used in the rational structure-based design of new PNP
inhibitors with potential medical applications.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Structures of the multisubstrate analogue inhibitor
DFPP-G and some of its precursors: non-fluorinated (DPP-G),
non-fluorinated and without terminal phosphonyl group (HPG) and
orthophosphate (P[i]).
|
 |
Figure 4.
Figure 4 Superposition of the active sites of three complexes of
calf spleen PNP with various inhibitors: (i) the binary complex
with the multisubstrate analogue inhibitor DFPP-G (cpk colours,
present structure; PDB code [254]1v48 ), (ii) the binary complex
with the multisubstrate analogue inhibitor (S)-PMPDAP (magenta;
PDB code [255]1lv8 , monomer A; Bzowska et al., 2004[256]
[Bzowska, A., Koellner, G., Wielgus-Kutrowska, B., Stroh, A.,
Raszewski, G., Hol�, A., Steiner, T. & Frank, J. (2004).
Submitted.]-[257][bluearr.gif] ) and (iii) the ternary complex
with the transition-state inhibitor immucillin G and phosphate
(green; PDB code [258]1b8n ; Kicska et al., 2002[259] [Kicska,
G. A., Tyler, P. C., Evans, G. B., Furneaux, R. H., Schramm, V.
L. & Kim, K. (2002). J. Biol. Chem. 277,
3226-3231.]-[260][bluearr.gif] ). Active-site amino acids that
form putative hydrogen bonds with the inhibitors are included
(see also Fig. 2[261] [link]-[262][turqarr.gif] ). In addition,
the location of Phe200 involved in [263][pi] -stacking
interaction with the inhibitor base is also shown (thin lines).
Phosphate and phosphonate groups of the multisubstrate analogue
inhibitors [DFPP-G and (S)-PMPDAP] and orthophosphate in the
ternary complex with immucillin G occupy the same position in
all three structures. The side chain of Arg84 in the complex
with (S)-PMPDAP (drawn in light magenta) is directed away from
the phosphate-binding site and does not form hydrogen bonds with
the phosphonate group of the inhibitor. Tyr88, Met219 and His257
only form direct hydrogen-bonding contacts in the complex with
immucillin G. The inhibition potency of the three inhibitors
shown in the figure spans almost six orders of magnitude: from
30 pM for immucillin G to 6 �M for (S)-PMPDAP (see text for
details). The number of direct hydrogen-bonding contacts formed
by these three ligands correlates well with their inhibition
potency as observed in solution.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2004,
60,
1417-1424)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Breer,
L.Glavas-Obrovac,
M.Suver,
S.Hikishima,
M.Hashimoto,
T.Yokomatsu,
B.Wielgus-Kutrowska,
L.Magnowska,
and
A.Bzowska
(2010).
9-Deazaguanine derivatives connected by a linker to difluoromethylene phosphonic acid are slow-binding picomolar inhibitors of trimeric purine nucleoside phosphorylase.
|
| |
FEBS J,
277,
1747-1760.
|
 |
|
|
|
|
 |
M.L.Bellows,
and
C.A.Floudas
(2010).
Computational methods for de novo protein design and its applications to the human immunodeficiency virus 1, purine nucleoside phosphorylase, ubiquitin specific protease 7, and histone demethylases.
|
| |
Curr Drug Targets,
11,
264-278.
|
 |
|
|
|
|
 |
B.J.Geiss,
A.A.Thompson,
A.J.Andrews,
R.L.Sons,
H.H.Gari,
S.M.Keenan,
and
O.B.Peersen
(2009).
Analysis of flavivirus NS5 methyltransferase cap binding.
|
| |
J Mol Biol,
385,
1643-1654.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

