 |
PDBsum entry 1v0h

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1v0h
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.11.1.11
- L-ascorbate peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ascorbate + H2O2 = L-dehydroascorbate + 2 H2O
|
 |
 |
 |
 |
 |
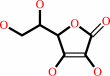 L-ascorbate
L-ascorbate
|
+
|
H2O2
Bound ligand (Het Group name = )
matches with 53.33% similarity
|
=
|
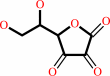 L-dehydroascorbate
L-dehydroascorbate
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
43:8644-8651
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the ascorbate peroxidase-salicylhydroxamic acid complex.
|
|
K.H.Sharp,
P.C.Moody,
K.A.Brown,
E.L.Raven.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ascorbate peroxidase is a bifunctional peroxidase that catalyzes the
H(2)O(2)-dependent oxidation of both ascorbate and various aromatic substrates.
The ascorbate binding site was recently identified as being close to the
gamma-heme edge [Sharp, K. H., Mewies, M., Moody, P. C. E., and Raven, E. L.
(2003)Nat. Struct. Biol. 10, 303-307]. In this work, the X-ray crystal structure
of recombinant soybean cytosolic ascorbate peroxidase (rsAPX) in complex with
salicylhydroxamic acid (SHA) has been determined to 1.46 A. The SHA molecule is
bound close to the delta-heme edge in a cavity that connects the distal side of
the heme to the surface of the protein. There are hydrogen bonds between the
phenolic hydroxide of the SHA and the main chain carbonyl of Pro132, between the
carbonyl oxygen of SHA and the side chain guanadinium group of Arg38, and
between the hydroxamic acid group and the indole nitrogen of Trp41. The
structure provides the first information about the location of the aromatic
binding site in ascorbate peroxidase and, together with our previous data
[Sharp, K. H., et al. (2003) Nat. Struct. Biol. 10, 303-307], completes the
structural description of the binding properties of ascorbate peroxidase. The
mechanistic implications of the results are discussed in terms of our current
understanding of how APX catalyzes oxidation of different types of substrates
bound at different locations.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Singh,
N.Singh,
M.Sinha,
A.Bhushan,
P.Kaur,
A.Srinivasan,
S.Sharma,
and
T.P.Singh
(2009).
Binding modes of aromatic ligands to mammalian heme peroxidases with associated functional implications: crystal structures of lactoperoxidase complexes with acetylsalicylic acid, salicylhydroxamic acid, and benzylhydroxamic acid.
|
| |
J Biol Chem,
284,
20311-20318.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Metcalfe,
I.K.Macdonald,
E.J.Murphy,
K.A.Brown,
E.L.Raven,
and
P.C.Moody
(2008).
The tuberculosis prodrug isoniazid bound to activating peroxidases.
|
| |
J Biol Chem,
283,
6193-6200.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Kitajima,
T.Shimaoka,
M.Kurioka,
and
A.Yokota
(2007).
Irreversible cross-linking of heme to the distal tryptophan of stromal ascorbate peroxidase in response to rapid inactivation by H2O2.
|
| |
FEBS J,
274,
3013-3020.
|
 |
|
|
|
|
 |
E.Gelhaye,
N.Navrot,
I.K.Macdonald,
N.Rouhier,
E.L.Raven,
and
J.P.Jacquot
(2006).
Ascorbate peroxidase-thioredoxin interaction.
|
| |
Photosynth Res,
89,
193-200.
|
 |
|
|
|
|
 |
S.K.Badyal,
M.G.Joyce,
K.H.Sharp,
H.E.Seward,
M.Mewies,
J.Basran,
I.K.Macdonald,
P.C.Moody,
and
E.L.Raven
(2006).
Conformational mobility in the active site of a heme peroxidase.
|
| |
J Biol Chem,
281,
24512-24520.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

