 |
PDBsum entry 1v08

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the zea maze beta-glucosidase-1 in complex with gluco-tetrazole
|
|
Structure:
|
 |
Beta-glucosidase. Chain: a, b. Synonym: chloroplast precursor, gentiobiase, cellobiase, beta-d- glucoside glucohydrolase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Zea mays. Organism_taxid: 4577. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.175
|
R-free:
|
0.205
|
|
|
Authors:
|
 |
J.Moriniere,L.Verdoucq,D.R.Bevan,A.Esen,B.Henrissat,M.Czjzek
|
Key ref:

|
 |
L.Verdoucq
et al.
(2004).
Structural determinants of substrate specificity in family 1 beta-glucosidases: novel insights from the crystal structure of sorghum dhurrinase-1, a plant beta-glucosidase with strict specificity, in complex with its natural substrate.
J Biol Chem,
279,
31796-31803.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
25-Mar-04
|
Release date:
|
20-May-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P49235
(HGGL1_MAIZE) -
4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside beta-D-glucosidase 1, chloroplastic from Zea mays
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
566 a.a.
491 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.2.1.182
- 4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
DIMBOA beta-D-glucoside + H2O = DIMBOA + D-glucose
|
|
2.
|
DIBOA beta-D-glucoside + H2O = DIBOA + D-glucose
|
|
 |
 |
 |
 |
 |
DIMBOA beta-D-glucoside
|
+
|
H2O
|
=
|
DIMBOA
Bound ligand (Het Group name = )
matches with 62.50% similarity
|
+
|
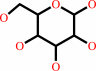 D-glucose
D-glucose
|
|
 |
 |
 |
 |
 |
DIBOA beta-D-glucoside
|
+
|
H2O
|
=
|
DIBOA
Bound ligand (Het Group name = )
matches with 62.50% similarity
|
+
|
 D-glucose
D-glucose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.2.1.21
- beta-glucosidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Hydrolysis of terminal, non-reducing beta-D-glucose residues with release of beta-D-glucose.
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:31796-31803
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural determinants of substrate specificity in family 1 beta-glucosidases: novel insights from the crystal structure of sorghum dhurrinase-1, a plant beta-glucosidase with strict specificity, in complex with its natural substrate.
|
|
L.Verdoucq,
J.Morinière,
D.R.Bevan,
A.Esen,
A.Vasella,
B.Henrissat,
M.Czjze.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Plant beta-glucosidases play a crucial role in defense against pests. They
cleave, with variable specificity, beta-glucosides to release toxic aglycone
moieties. The Sorghum bicolor beta-glucosidase isoenzyme Dhr1 has a strict
specificity for its natural substrate dhurrin
(p-hydroxy-(S)-mandelonitrile-beta-D-glucoside), whereas its close homolog, the
maize beta-glucosidase isoenzyme Glu1, which shares 72% sequence identity,
hydrolyzes a broad spectrum of substrates in addition to its natural substrate
2-O-beta-D-glucopyranosyl-4-hydroxy-7-methoxy-1,4-benzoxaxin-3-one. Structural
data from enzyme.substrate complexes of Dhr1 show that the mode of aglycone
binding differs from that previously observed in the homologous maize enzyme.
Specifically, the data suggest that Asn(259), Phe(261), and Ser(462), located in
the aglycone-binding site of S. bicolor Dhr1, are crucial for aglycone
recognition and binding. The tight binding of the aglycone moiety of dhurrin
promotes the stabilization of the reaction intermediate in which the glycone
moiety is in a deformed (1)S(3) conformation within the glycone-binding site,
ready for nucleophilic attack to occur. Compared with the broad specificity
maize beta-glucosidase, this different binding mode explains the narrow
specificity of sorghum dhurrinase-1.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
FIG. 4. Dhurrin bound in the active sites to SbDhr1-E189D
and glucotetrazole bound to ZmGlu1-E191D. A, electron density
surrounding the dhurrin molecule in the active site of
SbDhr1-E189D. The 2F[o] - F[c] Fourier difference maps at the
final stage of refinement are shown contoured at 1  above
the mean density. B, a slice in the surface representation of
SbDhr1 in complex with dhurrin showing the cyano group-binding
pocket. The dipole moment of the polar pocket, calculated with
GRASP (28), coincides with that of the cyano group. C, electron
density around the glucotetrazole molecule in the active site of
ZmGlu1-E191D. The F[o] - F[c] Fourier difference maps before
refinement are shown, calculated using only the enzyme model
phases without substrate, contoured at 2.5 above
the mean density. B, a slice in the surface representation of
SbDhr1 in complex with dhurrin showing the cyano group-binding
pocket. The dipole moment of the polar pocket, calculated with
GRASP (28), coincides with that of the cyano group. C, electron
density around the glucotetrazole molecule in the active site of
ZmGlu1-E191D. The F[o] - F[c] Fourier difference maps before
refinement are shown, calculated using only the enzyme model
phases without substrate, contoured at 2.5  above the mean density.
D, superimposition of the active sites of myrosinase (blue) and
ZmGlu1 (yellow), both in complex with the glucotetrazole
inhibitor molecule. above the mean density.
D, superimposition of the active sites of myrosinase (blue) and
ZmGlu1 (yellow), both in complex with the glucotetrazole
inhibitor molecule.
|
 |
Figure 5.
FIG. 5. Superimposition of the glucose moieties of
DIMBOA-Glc in ZmGlu1 and dhurrin in SbDhr1. The glucose ring of
DIM-BOA-Glc has rotated by  60° with respect to
that of dhurrin. Consequently, the residues binding the sugar
groups O-2, O-3, and O-4 in dhurrin bind O-3, O-4, and O-6 in
DIMBOA-Glc. See also Table III. In each box, the top residue
occurs in ZmGlu1, and the bottom residue occurs in SbDhr1. Of
the 2 glutamates shown by stick representation, Glu464 occurs in
ZmGlu1, whereas Glu460 occurs in SbDhr1. 60° with respect to
that of dhurrin. Consequently, the residues binding the sugar
groups O-2, O-3, and O-4 in dhurrin bind O-3, O-4, and O-6 in
DIMBOA-Glc. See also Table III. In each box, the top residue
occurs in ZmGlu1, and the bottom residue occurs in SbDhr1. Of
the 2 glutamates shown by stick representation, Glu464 occurs in
ZmGlu1, whereas Glu460 occurs in SbDhr1.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
31796-31803)
copyright 2004.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.R.Ketudat Cairns,
and
A.Esen
(2010).
β-Glucosidases.
|
| |
Cell Mol Life Sci,
67,
3389-3405.
|
 |
|
|
|
|
 |
A.D.Hill,
and
P.J.Reilly
(2008).
Computational analysis of glycoside hydrolase family 1 specificities.
|
| |
Biopolymers,
89,
1021-1031.
|
 |
|
|
|
|
 |
L.M.Mendonça,
and
S.R.Marana
(2008).
The role in the substrate specificity and catalysis of residues forming the substrate aglycone-binding site of a beta-glycosidase.
|
| |
FEBS J,
275,
2536-2547.
|
 |
|
|
|
|
 |
R.Dopitová,
P.Mazura,
L.Janda,
R.Chaloupková,
P.Jerábek,
J.Damborský,
T.Filipi,
N.S.Kiran,
and
B.Brzobohatý
(2008).
Functional analysis of the aglycone-binding site of the maize beta-glucosidase Zm-p60.1.
|
| |
FEBS J,
275,
6123-6135.
|
 |
|
|
|
|
 |
Z.Minic
(2008).
Physiological roles of plant glycoside hydrolases.
|
| |
Planta,
227,
723-740.
|
 |
|
|
|
|
 |
J.Stöckigt,
and
S.Panjikar
(2007).
Structural biology in plant natural product biosynthesis--architecture of enzymes from monoterpenoid indole and tropane alkaloid biosynthesis.
|
| |
Nat Prod Rep,
24,
1382-1400.
|
 |
|
|
|
|
 |
M.León,
P.Isorna,
M.Menéndez,
J.Sanz-Aparicio,
and
J.Polaina
(2007).
Comparative study and mutational analysis of distinctive structural elements of hyperthermophilic enzymes.
|
| |
Protein J,
26,
435-444.
|
 |
|
|
|
|
 |
B.Di Lauro,
M.Rossi,
and
M.Moracci
(2006).
Characterization of a beta-glycosidase from the thermoacidophilic bacterium Alicyclobacillus acidocaldarius.
|
| |
Extremophiles,
10,
301-310.
|
 |
|
|
|
|
 |
W.Chuenchor,
S.Pengthaisong,
J.Yuvaniyama,
R.Opassiri,
J.Svasti,
and
J.R.Ketudat Cairns
(2006).
Purification, crystallization and preliminary X-ray analysis of rice BGlu1 beta-glucosidase with and without 2-deoxy-2-fluoro-beta-D-glucoside.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
798-801.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

