 |
PDBsum entry 1rv8

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Class ii fructose-1,6-bisphosphate aldolase from thermus aquaticus in complex with cobalt
|
|
Structure:
|
 |
Fructose-1,6-bisphosphate aldolase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Thermus aquaticus. Organism_taxid: 271. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.211
|
R-free:
|
0.252
|
|
|
Authors:
|
 |
T.Izard,J.Sygusch
|
Key ref:

|
 |
T.Izard
and
J.Sygusch
(2004).
Induced fit movements and metal cofactor selectivity of class II aldolases: structure of Thermus aquaticus fructose-1,6-bisphosphate aldolase.
J Biol Chem,
279,
11825-11833.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
12-Dec-03
|
Release date:
|
27-Jan-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9RHA2
(Q9RHA2_THEAQ) -
Fructose-1,6-bisphosphate aldolase from Thermus aquaticus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
305 a.a.
297 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.2.13
- fructose-bisphosphate aldolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate = D-glyceraldehyde 3-phosphate + dihydroxyacetone phosphate
|
 |
 |
 |
 |
 |
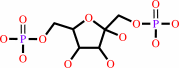 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
 dihydroxyacetone phosphate
dihydroxyacetone phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:11825-11833
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Induced fit movements and metal cofactor selectivity of class II aldolases: structure of Thermus aquaticus fructose-1,6-bisphosphate aldolase.
|
|
T.Izard,
J.Sygusch.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Fructose-1,6-bisphosphate (FBP) aldolase is an essential glycolytic enzyme that
reversibly cleaves its ketohexose substrate into triose phosphates. Here we
report the crystal structure of a metallo-dependent or class II FBP aldolase
from an extreme thermophile, Thermus aquaticus (Taq). The quaternary structure
reveals a tetramer composed of two dimers related by a 2-fold axis. Taq FBP
aldolase subunits exhibit two distinct conformational states corresponding to
loop regions that are in either open or closed position with respect to the
active site. Loop closure remodels the disposition of chelating active site
histidine residues. In subunits corresponding to the open conformation, the
metal cofactor, Co(2+), is sequestered in the active site, whereas for subunits
in the closed conformation, the metal cation exchanges between two mutually
exclusive binding loci, corresponding to a site at the active site surface and
an interior site vicinal to the metal-binding site in the open conformation.
Cofactor site exchange is mediated by rotations of the chelating histidine side
chains that are coupled to the prior conformational change of loop closure.
Sulfate anions are consistent with the location of the phosphate-binding sites
of the FBP substrate and determine not only the previously unknown second
phosphate-binding site but also provide a mechanism that regulates loop closure
during catalysis. Modeling of FBP substrate into the active site is consistent
with binding by the acyclic keto form, a minor solution species, and with the
metal cofactor mediating keto bond polarization. The Taq FBP aldolase structure
suggests a structural basis for different metal cofactor specificity than in
Escherichia coli FBP aldolase structures, and we discuss its potential role
during catalysis. Comparison with the E. coli structure also indicates a
structural basis for thermostability by Taq FBP aldolase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
FIG. 2. A cartoon drawing of the FBP aldolase oligomer with
point group 222. The three different molecular dyads comprise a
right-handed orthogonal set of axes P, Q, and R as originally
defined for the three 2-fold axes of lactate dehydrogenase (46).
In A, the view is looking down the crystallographic dyad (P),
while in B the orientation is looking down the molecular dyad
(R). The dyads (R and Q in A and P and Q in B) are indicated by
solid lines. Each protomer is shown in a different color.
|
 |
Figure 4.
FIG. 4. Stereo view of Taq FBP aldolase active site. Final
 [A]
weighted F[o] - F[c] omit electron density map for ligands bound
to the enzyme. The contour level of the electron density map is
4 [A]
weighted F[o] - F[c] omit electron density map for ligands bound
to the enzyme. The contour level of the electron density map is
4  ,
and the resolution is 2.3 Å. The bonds of the ligands are
drawn in pink, whereas the bonds of the enzyme are shown in
light gray. For clarity, water molecules (drawn as spheres) are
not labeled. Residues belonging to a 2-fold related subunit are
italicized. A, protomer in the closed conformation showing
residues in contact with the sulfate anions that coincide with
the phosphate-binding sites of FBP, the two mutually exclusive
Co2+ cofactors (drawn as light blue spheres) and the activating
cation (drawn as a black sphere). B, sulfate and cation binding
to the active site as observed in the subunits in their open
conformation. Orientation was rotated by 15° with respect to
A to reveal Asn251 that interacts with the monovalent cation. C,
FBP modeled into the active site using the sulfate-binding sites
of the closed protomer as phosphate oxyanion templates in the
Taq FBP aldolase complex with yttrium. The novel metal-binding
site (yttrium) is drawn as a green sphere. The hydroxyls O[2]
and O[3] of the FBP molecule are within close contact of the
exterior Co2+ site (indicated by dashed lines). ,
and the resolution is 2.3 Å. The bonds of the ligands are
drawn in pink, whereas the bonds of the enzyme are shown in
light gray. For clarity, water molecules (drawn as spheres) are
not labeled. Residues belonging to a 2-fold related subunit are
italicized. A, protomer in the closed conformation showing
residues in contact with the sulfate anions that coincide with
the phosphate-binding sites of FBP, the two mutually exclusive
Co2+ cofactors (drawn as light blue spheres) and the activating
cation (drawn as a black sphere). B, sulfate and cation binding
to the active site as observed in the subunits in their open
conformation. Orientation was rotated by 15° with respect to
A to reveal Asn251 that interacts with the monovalent cation. C,
FBP modeled into the active site using the sulfate-binding sites
of the closed protomer as phosphate oxyanion templates in the
Taq FBP aldolase complex with yttrium. The novel metal-binding
site (yttrium) is drawn as a green sphere. The hydroxyls O[2]
and O[3] of the FBP molecule are within close contact of the
exterior Co2+ site (indicated by dashed lines).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
11825-11833)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Galkin,
Z.Li,
L.Li,
L.Kulakova,
L.R.Pal,
D.Dunaway-Mariano,
and
O.Herzberg
(2009).
Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
|
| |
Biochemistry,
48,
3186-3196.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.D.Pegan,
K.Rukseree,
S.G.Franzblau,
and
A.D.Mesecar
(2009).
Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis.
|
| |
J Mol Biol,
386,
1038-1053.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.F.Waingeh,
C.D.Gustafson,
E.I.Kozliak,
S.L.Lowe,
H.R.Knull,
and
K.A.Thomasson
(2006).
Glycolytic enzyme interactions with yeast and skeletal muscle F-actin.
|
| |
Biophys J,
90,
1371-1384.
|
 |
|
|
|
|
 |
X.Li,
H.Huang,
X.Song,
Y.Wang,
H.Xu,
M.Teng,
and
W.Gong
(2006).
Purification, crystallization and preliminary crystallographic studies on 2-dehydro-3-deoxygalactarate aldolase from Leptospira interrogans.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1269-1270.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

