 |
PDBsum entry 1r4z

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.3
- triacylglycerol lipase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a triacylglycerol + H2O = a diacylglycerol + a fatty acid + H+
|
 |
 |
 |
 |
 |
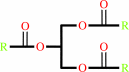 triacylglycerol
triacylglycerol
|
+
|
H2O
|
=
|
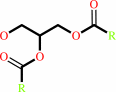 diacylglycerol
diacylglycerol
|
+
|
 fatty acid
fatty acid
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Chembiochem
7:149-157
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Directed evolution of Bacillus subtilis lipase A by use of enantiomeric phosphonate inhibitors: crystal structures and phage display selection.
|
|
M.J.Dröge,
Y.L.Boersma,
G.van Pouderoyen,
T.E.Vrenken,
C.J.Rüggeberg,
M.T.Reetz,
B.W.Dijkstra,
W.J.Quax.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phage display can be used as a protein-engineering tool for the selection of
proteins with desirable binding properties from a library of mutants. Here we
describe the application of this method for the directed evolution of Bacillus
subtilis lipase A, an enzyme that has important properties for the preparation
of the pharmaceutically relevant chiral compound
1,2-O-isopropylidene-sn-glycerol (IPG). PCR mutagenesis with spiked
oligonucleotides was employed for saturation mutagenesis of a stretch of amino
acids near the active site. After expression of these mutants on bacteriophages,
dual selection with (S)-(+)- and (R)-(-)-IPG stereoisomers covalently coupled to
enantiomeric phosphonate suicide inhibitors (SIRAN Sc and Rc inhibitors,
respectively) was used for the isolation of variants with inverted
enantioselectivity. The mutants were further characterised by determination of
their Michaelis-Menten parameters. The 3D structures of the Sc and Rc
inhibitor-lipase complexes were determined and provided structural insight into
the mechanism of enantioselectivity of the enzyme. In conclusion, we have used
phage display as a fast and reproducible method for the selection of Bacillus
lipase A mutant enzymes with inverted enantioselectivity.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.E.Cassimjee,
R.Kourist,
D.Lindberg,
M.Wittrup Larsen,
N.H.Thanh,
M.Widersten,
U.T.Bornscheuer,
and
P.Berglund
(2011).
One-step enzyme extraction and immobilization for biocatalysis applications.
|
| |
Biotechnol J,
6,
463-469.
|
 |
|
|
|
|
 |
Y.Jiang,
K.L.Morley,
J.D.Schrag,
and
R.J.Kazlauskas
(2011).
Different active-site loop orientation in serine hydrolases versus acyltransferases.
|
| |
Chembiochem,
12,
768-776.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Otte,
M.Bocola,
and
W.Thiel
(2009).
Force-field parameters for the simulation of tetrahedral intermediates of serine hydrolases.
|
| |
J Comput Chem,
30,
154-162.
|
 |
|
|
|
|
 |
Y.L.Boersma,
M.J.Dröge,
A.M.van der Sloot,
T.Pijning,
R.H.Cool,
B.W.Dijkstra,
and
W.J.Quax
(2008).
A novel genetic selection system for improved enantioselectivity of Bacillus subtilis lipase A.
|
| |
Chembiochem,
9,
1110-1115.
|
 |
|
|
|
|
 |
M.T.Reetz,
and
J.D.Carballeira
(2007).
Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes.
|
| |
Nat Protoc,
2,
891-903.
|
 |
|
|
|
|
 |
S.Becker,
A.Michalczyk,
S.Wilhelm,
K.E.Jaeger,
and
H.Kolmar
(2007).
Ultrahigh-throughput screening to identify E. coli cells expressing functionally active enzymes on their surface.
|
| |
Chembiochem,
8,
943-949.
|
 |
|
|
|
|
 |
Y.L.Boersma,
M.J.Dröge,
and
W.J.Quax
(2007).
Selection strategies for improved biocatalysts.
|
| |
FEBS J,
274,
2181-2195.
|
 |
|
|
|
|
 |
Z.Qian,
C.J.Fields,
Y.Yu,
and
S.Lutz
(2007).
Recent progress in engineering alpha/beta hydrolase-fold family members.
|
| |
Biotechnol J,
2,
192-200.
|
 |
|
|
|
|
 |
M.Konarzycka-Bessler,
and
K.E.Jaeger
(2006).
Select the best: novel biocatalysts for industrial applications.
|
| |
Trends Biotechnol,
24,
248-250.
|
 |
|
|
|
|
 |
M.T.Reetz,
J.D.Carballeira,
and
A.Vogel
(2006).
Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability.
|
| |
Angew Chem Int Ed Engl,
45,
7745-7751.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

