 |
PDBsum entry 1r3q

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.37
- uroporphyrinogen decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Porphyrin Biosynthesis (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
uroporphyrinogen III + 4 H+ = coproporphyrinogen III + 4 CO2
|
 |
 |
 |
 |
 |
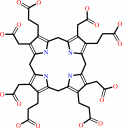 uroporphyrinogen III
uroporphyrinogen III
|
+
|
4
×
H(+)
|
=
|
coproporphyrinogen III
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
4
×
CO2
Bound ligand (Het Group name = )
matches with 84.62% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
22:6225-6233
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase.
|
|
J.D.Phillips,
F.G.Whitby,
J.P.Kushner,
C.P.Hill.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Uroporphyrinogen decarboxylase (URO-D), an essential enzyme that functions in
the heme biosynthetic pathway, catalyzes decarboxylation of all four acetate
groups of uroporphyrinogen to form coproporphyrinogen. Here we report crystal
structures of URO-D in complex with the I and III isomer coproporphyrinogen
products. Crystallization required use of a novel enzymatic approach to generate
the highly oxygen-sensitive porphyrinogen substrate in situ. The tetrapyrrole
product adopts a domed conformation that lies against a collar of conserved
hydrophobic residues and allows formation of hydrogen bonding interactions
between a carboxylate oxygen atom of the invariant Asp86 residue and the pyrrole
NH groups. Structural and biochemical analyses of URO-D proteins mutated at
Asp86 support the conclusion that this residue makes important contributions to
binding and likely promotes catalysis by stabilizing a positive charge on a
reaction intermediate. The central coordination geometry of Asp86 allows the
initial substrates and the various partially decarboxylated intermediates to be
bound with equivalent activating interactions, and thereby explains how all four
of the substrate acetate groups can be decarboxylated at the same catalytic
center.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Reaction catalyzed by URO-D and the two preceding steps
in the heme biosynthesis pathway. Acetate, propionate and methyl
side-chains are denoted A, P and M, respectively.
Porphobilinogen deaminase (PBG-D), uroporphyrinogen III synthase
(U3S). In the absence of U3S, hydroxymethylbilane cyclizes
without inversion of the D-ring, to form uroporphyrinogen I,
which differs from the III-isomer shown here by having an
identical arrangement of A/P substituents on all four pyrrole
rings. Figures 1 and 4 were generated using ChemDraw Pro
(CambridgeSoft Corp., Cambridge, MA).
|
 |
Figure 3.
Figure 3 Interactions at the active site. (A) Uroporphyrinogen I
product (green), wild-type URO-D (yellow). Protein residues are
shown if one atom from the residue lies within 4.0 Å of the
product in at least one of the structures. Also shown is Leu88.
The pyrrole rings are denoted A, B, C and D, with the D-ring
being the site where acetate and propionate groups are reversed
in the III-isomer product. Van der Waals' surfaces are shown
around the protein atoms, with residues nearer the viewer given
a more transparent surface. Hydrogen bonds are indicated with
dashed lines. The apparent hydrogen bond seen between Ala39 O
and the C-ring propionate indicates that this carboxylate is
protonated. (B) Same as (A), but for the III-isomer product
complex. The I- and III-isomer products superimpose very closely
following overlap on the protein C[  ]atoms.
The major differences are the conformations of Arg37 and Arg 41
side chains. (C) Comparison of product bound to various URO-D
variants. Structures were superimposed by overlap on the protein
C[ ]atoms.
The major differences are the conformations of Arg37 and Arg 41
side chains. (C) Comparison of product bound to various URO-D
variants. Structures were superimposed by overlap on the protein
C[  ]atoms.
The water molecules that lie roughly in the position of the
wild-type Asp86 side chains are shown explicitly. Color scheme
is indicated and is the same as in Figure 2B. This figure was
generated using Molscript (Kraulis, 1991) and Raster3D (Merritt
and Bacon, 1997). ]atoms.
The water molecules that lie roughly in the position of the
wild-type Asp86 side chains are shown explicitly. Color scheme
is indicated and is the same as in Figure 2B. This figure was
generated using Molscript (Kraulis, 1991) and Raster3D (Merritt
and Bacon, 1997).
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(2003,
22,
6225-6233)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.A.Bushnell,
E.Erdtman,
J.Llano,
L.A.Eriksson,
and
J.W.Gauld
(2011).
The first branching point in porphyrin biosynthesis: A systematic docking, molecular dynamics and quantum mechanical/molecular mechanical study of substrate binding and mechanism of uroporphyrinogen-III decarboxylase.
|
| |
J Comput Chem,
32,
822-834.
|
 |
|
|
|
|
 |
B.C.Tripathy,
I.Sherameti,
and
R.Oelmüller
(2010).
Siroheme: an essential component for life on earth.
|
| |
Plant Signal Behav,
5,
14-20.
|
 |
|
|
|
|
 |
C.Ng,
M.Z.DeMaere,
T.J.Williams,
F.M.Lauro,
M.Raftery,
J.A.Gibson,
C.Andrews-Pfannkoch,
M.Lewis,
J.M.Hoffman,
T.Thomas,
and
R.Cavicchioli
(2010).
Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica.
|
| |
ISME J,
4,
1002-1019.
|
 |
|
|
|
|
 |
G.Layer,
J.Reichelt,
D.Jahn,
and
D.W.Heinz
(2010).
Structure and function of enzymes in heme biosynthesis.
|
| |
Protein Sci,
19,
1137-1161.
|
 |
|
|
|
|
 |
C.A.Warby,
J.D.Phillips,
H.A.Bergonia,
F.G.Whitby,
C.P.Hill,
and
J.P.Kushner
(2009).
Structural and kinetic characterization of mutant human uroporphyrinogen decarboxylases.
|
| |
Cell Mol Biol (Noisy-le-grand),
55,
40-45.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.A.Bergonia,
J.D.Phillips,
and
J.P.Kushner
(2009).
Reduction of porphyrins to porphyrinogens with palladium on carbon.
|
| |
Anal Biochem,
384,
74-78.
|
 |
|
|
|
|
 |
J.D.Phillips,
C.A.Warby,
F.G.Whitby,
J.P.Kushner,
and
C.P.Hill
(2009).
Substrate shuttling between active sites of uroporphyrinogen decarboxylase is not required to generate coproporphyrinogen.
|
| |
J Mol Biol,
389,
306-314.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Masoumi,
I.U.Heinemann,
M.Rohde,
M.Koch,
M.Jahn,
and
D.Jahn
(2008).
Complex formation between protoporphyrinogen IX oxidase and ferrochelatase during haem biosynthesis in Thermosynechococcus elongatus.
|
| |
Microbiology,
154,
3707-3714.
|
 |
|
|
|
|
 |
C.A.Lewis,
and
R.Wolfenden
(2008).
Uroporphyrinogen decarboxylation as a benchmark for the catalytic proficiency of enzymes.
|
| |
Proc Natl Acad Sci U S A,
105,
17328-17333.
|
 |
|
|
|
|
 |
H.L.Schubert,
J.D.Phillips,
A.Heroux,
and
C.P.Hill
(2008).
Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex.
|
| |
Biochemistry,
47,
8648-8655.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Medlock,
L.Swartz,
T.A.Dailey,
H.A.Dailey,
and
W.N.Lanzilotta
(2007).
Substrate interactions with human ferrochelatase.
|
| |
Proc Natl Acad Sci U S A,
104,
1789-1793.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.D.Phillips,
H.A.Bergonia,
C.A.Reilly,
M.R.Franklin,
and
J.P.Kushner
(2007).
A porphomethene inhibitor of uroporphyrinogen decarboxylase causes porphyria cutanea tarda.
|
| |
Proc Natl Acad Sci U S A,
104,
5079-5084.
|
 |
|
|
|
|
 |
J.R.Stephenson,
J.A.Stacey,
J.B.Morgenthaler,
J.A.Friesen,
T.D.Lash,
and
M.A.Jones
(2007).
Role of aspartate 400, arginine 262, and arginine 401 in the catalytic mechanism of human coproporphyrinogen oxidase.
|
| |
Protein Sci,
16,
401-410.
|
 |
|
|
|
|
 |
M.Méndez,
P.Poblete-Gutiérrez,
M.García-Bravo,
T.Wiederholt,
M.J.Morán-Jiménez,
H.F.Merk,
M.C.Garrido-Astray,
J.Frank,
A.Fontanellas,
and
R.Enríquez de Salamanca
(2007).
Molecular heterogeneity of familial porphyria cutanea tarda in Spain: characterization of 10 novel mutations in the UROD gene.
|
| |
Br J Dermatol,
157,
501-507.
|
 |
|
|
|
|
 |
G.Layer,
A.J.Pierik,
M.Trost,
S.E.Rigby,
H.K.Leech,
K.Grage,
D.Breckau,
I.Astner,
L.Jänsch,
P.Heathcote,
M.J.Warren,
D.W.Heinz,
and
D.Jahn
(2006).
The substrate radical of Escherichia coli oxygen-independent coproporphyrinogen III oxidase HemN.
|
| |
J Biol Chem,
281,
15727-15734.
|
 |
|
|
|
|
 |
S.Al-Karadaghi,
R.Franco,
M.Hansson,
J.A.Shelnutt,
G.Isaya,
and
G.C.Ferreira
(2006).
Chelatases: distort to select?
|
| |
Trends Biochem Sci,
31,
135-142.
|
 |
|
|
|
|
 |
E.Raux-Deery,
H.K.Leech,
K.A.Nakrieko,
K.J.McLean,
A.W.Munro,
P.Heathcote,
S.E.Rigby,
A.G.Smith,
and
M.J.Warren
(2005).
Identification and characterization of the terminal enzyme of siroheme biosynthesis from Arabidopsis thaliana: a plastid-located sirohydrochlorin ferrochelatase containing a 2FE-2S center.
|
| |
J Biol Chem,
280,
4713-4721.
|
 |
|
|
|
|
 |
G.Chaufan,
M.M.Corvi,
L.C.San Martín de Viale,
M.L.Cárdenas,
and
M.d.e.l. .C.Ríos de Molina
(2005).
Abnormal kinetic behavior of uroporphyrinogen decarboxylase obtained from rats with hexachlorobenzene-induced porphyria.
|
| |
J Biochem Mol Toxicol,
19,
19-24.
|
 |
|
|
|
|
 |
G.Layer,
K.Grage,
T.Teschner,
V.Schünemann,
D.Breckau,
A.Masoumi,
M.Jahn,
P.Heathcote,
A.X.Trautwein,
and
D.Jahn
(2005).
Radical S-adenosylmethionine enzyme coproporphyrinogen III oxidase HemN: functional features of the [4Fe-4S] cluster and the two bound S-adenosyl-L-methionines.
|
| |
J Biol Chem,
280,
29038-29046.
|
 |
|
|
|
|
 |
J.D.Phillips,
F.G.Whitby,
C.A.Warby,
P.Labbe,
C.Yang,
J.W.Pflugrath,
J.D.Ferrara,
H.Robinson,
J.P.Kushner,
and
C.P.Hill
(2004).
Crystal structure of the oxygen-dependant coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae.
|
| |
J Biol Chem,
279,
38960-38968.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

