 |
PDBsum entry 1qw7

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.8.1
- aryldialkylphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
An aryl dialkyl phosphate + H2O = dialkyl phosphate + an aryl alcohol
|
 |
 |
 |
 |
 |
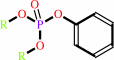 aryl dialkyl phosphate
aryl dialkyl phosphate
|
+
|
H2O
|
=
|
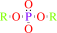 dialkyl phosphate
dialkyl phosphate
|
+
|
aryl alcohol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Arch Biochem Biophys
442:169-179
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and mutational studies of organophosphorus hydrolase reveal a cryptic and functional allosteric-binding site.
|
|
J.K.Grimsley,
B.Calamini,
J.R.Wild,
A.D.Mesecar.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Organophosphorus hydrolase detoxifies a broad range of organophosphate
pesticides and the chemical warfare agents (CWAs) sarin and VX. Previously,
rational genetic engineering produced OPH variants with 30-fold enhancements in
the hydrolysis of CWA and their analogs. One interesting variant (H254R) in
which the histidine at position 254 was changed to an arginine showed a 4-fold
increase in the hydrolysis of demetonS (VX analog), a 14-fold decrease with
paraoxon (an insecticide), and a 183-fold decrease with DFP (sarin analog). The
three-dimensional structure of this enzyme at 1.9A resolution with the
inhibitor, diethyl 4-methylbenzylphosphonate (EBP), revealed that the inhibitor
did not bind at the active site, but bound exclusively into a well-defined
surface pocket 12 A away from the active site. This structural feature was
accompanied by non-competitive inhibition of paraoxon hydrolysis by EBP with
H254R, in contrast to the native enzyme, which showed competitive inhibition.
These parallel structure-function characteristics identify a functional,
allosteric site on the surface of this enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.K.Raynes,
F.G.Pearce,
S.J.Meade,
and
J.A.Gerrard
(2011).
Immobilization of organophosphate hydrolase on an amyloid fibril nanoscaffold: Towards bioremediation and chemical detoxification.
|
| |
Biotechnol Prog,
27,
360-367.
|
 |
|
|
|
|
 |
C.Gilley,
M.MacDonald,
F.Nachon,
L.M.Schopfer,
J.Zhang,
J.R.Cashman,
and
O.Lockridge
(2009).
Nerve agent analogues that produce authentic soman, sarin, tabun, and cyclohexyl methylphosphonate-modified human butyrylcholinesterase.
|
| |
Chem Res Toxicol,
22,
1680-1688.
|
 |
|
|
|
|
 |
C.J.Jackson,
J.L.Foo,
N.Tokuriki,
L.Afriat,
P.D.Carr,
H.K.Kim,
G.Schenk,
D.S.Tawfik,
and
D.L.Ollis
(2009).
Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase.
|
| |
Proc Natl Acad Sci U S A,
106,
21631-21636.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Zhang,
R.Wu,
L.Song,
Y.Lin,
M.Lin,
Z.Cao,
W.Wu,
and
Y.Mo
(2009).
Molecular dynamics simulations of the detoxification of paraoxon catalyzed by phosphotriesterase.
|
| |
J Comput Chem,
30,
2388-2401.
|
 |
|
|
|
|
 |
C.D.Fleming,
C.C.Edwards,
S.D.Kirby,
D.M.Maxwell,
P.M.Potter,
D.M.Cerasoli,
and
M.R.Redinbo
(2007).
Crystal structures of human carboxylesterase 1 in covalent complexes with the chemical warfare agents soman and tabun.
|
| |
Biochemistry,
46,
5063-5071.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Ramanathan,
and
A.L.Simonian
(2007).
Array biosensor based on enzyme kinetics monitoring by fluorescence spectroscopy: application for neurotoxins detection.
|
| |
Biosens Bioelectron,
22,
3001-3007.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

