 |
PDBsum entry 1ps8

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ps8
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.2.1.11
- aspartate-semialdehyde dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Lysine biosynthesis (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate 4-semialdehyde + phosphate + NADP+ = 4-phospho-L-aspartate + NADPH + H+
|
 |
 |
 |
 |
 |
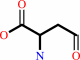 L-aspartate 4-semialdehyde
L-aspartate 4-semialdehyde
|
+
|
 phosphate
phosphate
|
+
|
 NADP(+)
NADP(+)
|
=
|
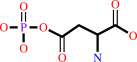 4-phospho-L-aspartate
4-phospho-L-aspartate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
60:1388-1395
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
|
|
J.Blanco,
R.A.Moore,
C.R.Faehnle,
D.M.Coe,
R.E.Viola.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The reversible dephosphorylation of beta-aspartyl phosphate to
L-aspartate-beta-semialdehyde (ASA) in the aspartate biosynthetic pathway is
catalyzed by aspartate-beta-semialdehyde dehydrogenase (ASADH). The product of
this reaction is a key intermediate in the biosynthesis of diaminopimelic acid,
an integral component of bacterial cell walls and a metabolic precursor of
lysine and also a precursor in the biosynthesis of threonine, isoleucine and
methionine. The structures of selected Haemophilus influenzae ASADH mutants were
determined in order to evaluate the residues that are proposed to interact with
the substrates ASA or phosphate. The substrate Km values are not altered by
replacement of either an active-site arginine (Arg270) with a lysine or a
putative phosphate-binding group (Lys246) with an arginine. However, the
interaction of phosphate with the enzyme is adversely affected by replacement of
Arg103 with lysine and is significantly altered when a neutral leucine is
substituted at this position. A conservative Glu243 to aspartate mutant does not
alter either ASA or phosphate binding, but instead results in an eightfold
increase in the Km for the coenzyme NADP. Each of the mutations is shown to
cause specific subtle active-site structural alterations and each of these
changes results in decreases in catalytic efficiency ranging from significant
(approximately 3% native activity) to substantial (<0.1% native activity).

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 An abbreviated mechanism of the reaction catalyzed by
aspartate  -semialdehyde
dehydrogenase. -semialdehyde
dehydrogenase.
|
 |
Figure 4.
Figure 4 Overlay of the backbone drawings of native (light) and
R103L mutant (dark) hiASADH structures. (a) The overall fold and
backbone position of the native and mutant structures are
essentially the same, except for the reorientation of a critical
active-site loop (shown in red). (b) Disruption of the
water-mediated hydrogen-bonding network between Arg103 and
Asn135 in the R103L mutant that triggers this loop movement and
shifts the position of the active-site Cys136 nucleophile. The
calculation for the overlay was performed with XtalView and the
drawing was produced using SPOCK.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2004,
60,
1388-1395)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Singh,
H.R.Kushwaha,
and
P.Sharma
(2008).
Molecular modelling and comparative structural account of aspartyl beta-semialdehyde dehydrogenase of Mycobacterium tuberculosis (H37Rv).
|
| |
J Mol Model,
14,
249-263.
|
 |
|
|
|
|
 |
T.Nonaka,
A.Kita,
J.Miura-Ohnuma,
E.Katoh,
N.Inagaki,
T.Yamazaki,
and
K.Miki
(2005).
Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa).
|
| |
Proteins,
61,
1137-1140.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

