 |
PDBsum entry 1nir

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Nitrite reductase
|
PDB id
|
|

|

|
1nir
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.7.2.1
- nitrite reductase (NO-forming).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
nitric oxide + Fe(III)-[cytochrome c] + H2O = Fe(II)-[cytochrome c] + nitrite + 2 H+
|
 |
 |
 |
 |
 |
 nitric oxide
nitric oxide
|
+
|
Fe(III)-[cytochrome c]
|
+
|
H2O
|
=
|
Fe(II)-[cytochrome c]
|
+
|
 nitrite
nitrite
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation or Fe cation; FAD
|
 |
 |
 |
 |
 |
Cu cation
|
or
|
Fe cation
|
 FAD
FAD
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.1.7.99.1
- hydroxylamine reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
A + NH4+ + H2O = hydroxylamine + AH2 + H+
|
 |
 |
 |
 |
 |
|
+
|
NH4(+)
|
+
|
H2O
|
=
|
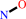 hydroxylamine
hydroxylamine
|
+
|
AH2
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Flavoprotein
|
 |
 |
 |
 |
 |
 Iron-sulfur
Iron-sulfur
|
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
5:1157-1171
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
N-terminal arm exchange is observed in the 2.15 A crystal structure of oxidized nitrite reductase from Pseudomonas aeruginosa.
|
|
D.Nurizzo,
M.C.Silvestrini,
M.Mathieu,
F.Cutruzzolà,
D.Bourgeois,
V.Fülöp,
J.Hajdu,
M.Brunori,
M.Tegoni,
C.Cambillau.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Nitrite reductase from Pseudomonas aeruginosa (NiR-Pa) is a dimer
consisting of two identical 60 kDa subunits, each of which contains one c and
one d1 heme group. This enzyme, a soluble component of the electron-transfer
chain that uses nitrate as a source of energy, can be induced by the addition of
nitrate to the bacterial growth medium. NiR-Pa catalyzes the reduction of
nitrite (NO2-) to nitric oxide (NO); in vitro, both cytochrome c551 and azurin
are efficient electron donors in this reaction. NiR is a key denitrification
enzyme, which controls the rate of the production of toxic nitric oxide (NO) and
ultimately regulates the release of NO into the atmosphere. RESULTS: The
structure of the orthorhombic form (P2(1)2(1)2) of oxidized NiR-Pa was solved at
2.15 A resolution, using molecular replacement with the coordinates of the NiR
from Thiosphaera pantotropha (NiR-Tp) as the starting model. Although the
d1-heme domains are almost identical in both enzyme structures, the c domain of
NiR-Pa is more like the classical class I cytochrome-c fold because it has His51
and Met88 as heme ligands, instead of His17 and His69 present in NiR-Tp. In
addition, the methionine-bearing loop, which was displaced by His17 of the
NiR-Tp N-terminal segment, is back to normal in our structure. The N-terminal
residues (5/6-30) of NiR-Pa and NiR-Tp have little sequence identity. In Nir-Pa,
this N-terminal segment of one monomer crosses the dimer interface and wraps
itself around the other monomer. Tyr10 of this segment is hydrogen bonded to an
hydroxide ion--the sixth ligand of the d1-heme Fe, whereas the equivalent
residue in NiR-Tp, Tyr25, is directly bound to the Fe. CONCLUSIONS: Two ligands
of hemes c and d1 differ between the two known NiR structures, which accounts
for the fact that they have quite different spectroscopic and kinetic features.
The unexpected domain-crossing by the N-terminal segment of NiR-Pa is comparable
to that of 'domain swapping' or 'arm exchange' previously observed in other
systems and may explain the observed cooperativity between monomers of dimeric
NiR-Pa. In spite of having similar sequence and fold, the different kinetic
behaviour and the spectral features of NiR-Pa and NiR-Tp are tuned by the
N-terminal stretch of residues. A further example of this may come from another
NiR, from Pseudomonas stutzeri, which has an N terminus very different from that
of the two above mentioned NiRs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 6.
Figure 6. Water-accessible surface area of NiR-Pa monomer
A, slabbed at the level of the d[1] heme. Water molecules are
colored in blue, the d[1] heme in red and the phosphate ion in
green. The channel starting from the d[1] heme and leading to
the `back door' is located between the d[1] heme and the
phosphate ion.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1997,
5,
1157-1171)
copyright 1997.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Rinaldo,
G.Giardina,
N.Castiglione,
V.Stelitano,
and
F.Cutruzzolà
(2011).
The catalytic mechanism of Pseudomonas aeruginosa cd1 nitrite reductase.
|
| |
Biochem Soc Trans,
39,
195-200.
|
 |
|
|
|
|
 |
F.Cutruzzolà,
S.Rinaldo,
N.Castiglione,
G.Giardina,
I.Pecht,
and
M.Brunori
(2009).
Nitrite reduction: a ubiquitous function from a pre-aerobic past.
|
| |
Bioessays,
31,
885-891.
|
 |
|
|
|
|
 |
O.Farver,
M.Brunori,
F.Cutruzzolà,
S.Rinaldo,
S.Wherland,
and
I.Pecht
(2009).
Intramolecular electron transfer in Pseudomonas aeruginosa cd(1) nitrite reductase: thermodynamics and kinetics.
|
| |
Biophys J,
96,
2849-2856.
|
 |
|
|
|
|
 |
S.Brenner,
D.J.Heyes,
S.Hay,
M.A.Hough,
R.R.Eady,
S.S.Hasnain,
and
N.S.Scrutton
(2009).
Demonstration of proton-coupled electron transfer in the copper-containing nitrite reductases.
|
| |
J Biol Chem,
284,
25973-25983.
|
 |
|
|
|
|
 |
S.Ghosh,
A.Dey,
Y.Sun,
C.P.Scholes,
and
E.I.Solomon
(2009).
Spectroscopic and computational studies of nitrite reductase: proton induced electron transfer and backbonding contributions to reactivity.
|
| |
J Am Chem Soc,
131,
277-288.
|
 |
|
|
|
|
 |
F.Jacobson,
A.Pistorius,
D.Farkas,
W.De Grip,
O.Hansson,
L.Sjölin,
and
R.Neutze
(2007).
pH dependence of copper geometry, reduction potential, and nitrite affinity in nitrite reductase.
|
| |
J Biol Chem,
282,
6347-6355.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Jacobson,
H.Guo,
K.Olesen,
M.Okvist,
R.Neutze,
and
L.Sjölin
(2005).
Structures of the oxidized and reduced forms of nitrite reductase from Rhodobacter sphaeroides 2.4.3 at high pH: changes in the interactions of the type 2 copper.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1190-1198.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.S.Zajicek,
M.R.Cheesman,
E.H.Gordon,
and
S.J.Ferguson
(2005).
Y25S variant of Paracoccus pantotrophus cytochrome cd1 provides insight into anion binding by d1 heme and a rare example of a critical difference between solution and crystal structures.
|
| |
J Biol Chem,
280,
26073-26079.
|
 |
|
|
|
|
 |
O.Einsle,
and
P.M.Kroneck
(2004).
Structural basis of denitrification.
|
| |
Biol Chem,
385,
875-883.
|
 |
|
|
|
|
 |
E.H.Gordon,
T.Sjögren,
M.Löfqvist,
C.D.Richter,
J.W.Allen,
C.W.Higham,
J.Hajdu,
V.Fülöp,
and
S.J.Ferguson
(2003).
Structure and kinetic properties of Paracoccus pantotrophus cytochrome cd1 nitrite reductase with the d1 heme active site ligand tyrosine 25 replaced by serine.
|
| |
J Biol Chem,
278,
11773-11781.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.D.Richter,
J.W.Allen,
C.W.Higham,
A.Koppenhofer,
R.S.Zajicek,
N.J.Watmough,
and
S.J.Ferguson
(2002).
Cytochrome cd1, reductive activation and kinetic analysis of a multifunctional respiratory enzyme.
|
| |
J Biol Chem,
277,
3093-3100.
|
 |
|
|
|
|
 |
F.Cutruzzola,
K.Brown,
E.K.Wilson,
A.Bellelli,
M.Arese,
M.Tegoni,
C.Cambillau,
and
M.Brunori
(2001).
The nitrite reductase from Pseudomonas aeruginosa: essential role of two active-site histidines in the catalytic and structural properties.
|
| |
Proc Natl Acad Sci U S A,
98,
2232-2237.
|
 |
|
|
|
|
 |
I.Moura,
and
J.J.Moura
(2001).
Structural aspects of denitrifying enzymes.
|
| |
Curr Opin Chem Biol,
5,
168-175.
|
 |
|
|
|
|
 |
K.Kobayashi,
A.Koppenhöfer,
S.J.Ferguson,
N.J.Watmough,
and
S.Tagawa
(2001).
Intramolecular electron transfer from c heme to d1 heme in bacterial cytochrome cd1 nitrite reductase occurs over the same distances at very different rates depending on the source of the enzyme.
|
| |
Biochemistry,
40,
8542-8547.
|
 |
|
|
|
|
 |
A.Koppenhöfer,
K.L.Turner,
J.W.Allen,
S.K.Chapman,
and
S.J.Ferguson
(2000).
Cytochrome cd(1) from Paracoccus pantotrophus exhibits kinetically gated, conformationally dependent, highly cooperative two-electron redox behavior.
|
| |
Biochemistry,
39,
4243-4249.
|
 |
|
|
|
|
 |
A.Koppenhöfer,
R.H.Little,
D.J.Lowe,
S.J.Ferguson,
and
N.J.Watmough
(2000).
Oxidase reaction of cytochrome cd(1) from Paracoccus pantotrophus.
|
| |
Biochemistry,
39,
4028-4036.
|
 |
|
|
|
|
 |
C.G.Friedrich,
A.Quentmeier,
F.Bardischewsky,
D.Rother,
R.Kraft,
S.Kostka,
and
H.Prinz
(2000).
Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17.
|
| |
J Bacteriol,
182,
4677-4687.
|
 |
|
|
|
|
 |
G.Ranghino,
E.Scorza,
T.Sjögren,
P.A.Williams,
M.Ricci,
and
J.Hajdu
(2000).
Quantum mechanical interpretation of nitrite reduction by cytochrome cd1 nitrite reductase from Paracoccus pantotrophus.
|
| |
Biochemistry,
39,
10958-10966.
|
 |
|
|
|
|
 |
T.Sjögren,
M.Svensson-Ek,
J.Hajdu,
and
P.Brzezinski
(2000).
Proton-coupled structural changes upon binding of carbon monoxide to cytochrome cd1: a combined flash photolysis and X-ray crystallography study.
|
| |
Biochemistry,
39,
10967-10974.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.R.Crane,
R.J.Rosenfeld,
A.S.Arvai,
D.K.Ghosh,
S.Ghosh,
J.A.Tainer,
D.J.Stuehr,
and
E.D.Getzoff
(1999).
N-terminal domain swapping and metal ion binding in nitric oxide synthase dimerization.
|
| |
EMBO J,
18,
6271-6281.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.J.Richardson,
and
N.J.Watmough
(1999).
Inorganic nitrogen metabolism in bacteria.
|
| |
Curr Opin Chem Biol,
3,
207-219.
|
 |
|
|
|
|
 |
D.K.Ghosh,
B.R.Crane,
S.Ghosh,
D.Wolan,
R.Gachhui,
C.Crooks,
A.Presta,
J.A.Tainer,
E.D.Getzoff,
and
D.J.Stuehr
(1999).
Inducible nitric oxide synthase: role of the N-terminal beta-hairpin hook and pterin-binding segment in dimerization and tetrahydrobiopterin interaction.
|
| |
EMBO J,
18,
6260-6270.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Nurizzo,
F.Cutruzzolà,
M.Arese,
D.Bourgeois,
M.Brunori,
C.Cambillau,
and
M.Tegoni
(1999).
Does the reduction of c heme trigger the conformational change of crystalline nitrite reductase?
|
| |
J Biol Chem,
274,
14997-15004.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.K.Wilson,
A.Bellelli,
S.Liberti,
M.Arese,
S.Grasso,
F.Cutruzzolà,
M.Brunori,
and
P.Brzezinski
(1999).
Internal electron transfer and structural dynamics of cd1 nitrite reductase revealed by laser CO photodissociation.
|
| |
Biochemistry,
38,
7556-7564.
|
 |
|
|
|
|
 |
F.Cutruzzolà
(1999).
Bacterial nitric oxide synthesis.
|
| |
Biochim Biophys Acta,
1411,
231-249.
|
 |
|
|
|
|
 |
D.Nurizzo,
F.Cutruzzolà,
M.Arese,
D.Bourgeois,
M.Brunori,
C.Cambillau,
and
M.Tegoni
(1998).
Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
|
| |
Biochemistry,
37,
13987-13996.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.J.Ferguson
(1998).
Nitrogen cycle enzymology.
|
| |
Curr Opin Chem Biol,
2,
182-193.
|
 |
|
|
|
|
 |
S.Wang,
W.R.Trumble,
H.Liao,
C.R.Wesson,
A.K.Dunker,
and
C.H.Kang
(1998).
Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum.
|
| |
Nat Struct Biol,
5,
476-483.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

