 |
PDBsum entry 1n4w

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1n4w
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.3.6
- cholesterol oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
cholesterol + O2 = cholest-5-en-3-one + H2O2
|
 |
 |
 |
 |
 |
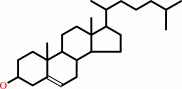 cholesterol
cholesterol
|
+
|
 O2
O2
|
=
|
cholest-5-en-3-one
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.5.3.3.1
- steroid Delta-isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 3-oxo-Delta5-steroid = a 3-oxo-Delta4-steroid
|
 |
 |
 |
 |
 |
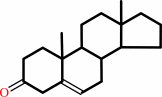 3-oxo-Delta(5)-steroid
3-oxo-Delta(5)-steroid
|
=
|
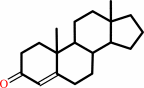 3-oxo-Delta(4)-steroid
3-oxo-Delta(4)-steroid
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Chem Biol
2:259-264
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Atomic resolution crystallography reveals how changes in pH shape the protein microenvironment.
|
|
A.Y.Lyubimov,
P.I.Lario,
I.Moustafa,
A.Vrielink.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Hydrogen atoms are a vital component of enzyme structure and function. In recent
years, atomic resolution crystallography (>or=1.2 A) has been successfully used
to investigate the role of the hydrogen atom in enzymatic catalysis. Here,
atomic resolution crystallography was used to study the effect of pH on
cholesterol oxidase from Streptomyces sp., a flavoenzyme oxidoreductase.
Crystallographic observations of the anionic oxidized flavin cofactor at basic
pH are consistent with the UV-visible absorption profile of the enzyme and
readily explain the reversible pH-dependent loss of oxidation activity.
Furthermore, a hydrogen atom, positioned at an unusually short distance from the
main chain carbonyl oxygen of Met122 at high pH, was observed, suggesting a
previously unknown mechanism of cofactor stabilization. This study shows how a
redox active site responds to changes in the enzyme's environment and how these
changes are able to influence the mechanism of enzymatic catalysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
(a–e) Electron-density features around the imidazole ring
of His447 at pH 4.5 (a), pH 5.2 (b), pH 5.8 (c), pH 7.3 (d) and
pH 9.0 (e). (f) A view of the interactions between His447,
Asn321 and Asn323 at pH 5.2. The 2F[o] – F[c] density
(magenta) is contoured at 4.0  ,
and the sharpened F[o] – F[c] density (green) is contoured at
2.0 ,
and the sharpened F[o] – F[c] density (green) is contoured at
2.0  .
The atoms are depicted by a ball-and-stick representation. The
bifurcated hydrogen bonds formed by ND1 of His447 with the amide
groups of Asn321 and Asn323 are drawn as blue dashed lines. The
chemical structure of the histidine side chain with labeled
atoms is included. .
The atoms are depicted by a ball-and-stick representation. The
bifurcated hydrogen bonds formed by ND1 of His447 with the amide
groups of Asn321 and Asn323 are drawn as blue dashed lines. The
chemical structure of the histidine side chain with labeled
atoms is included.
|
 |
Figure 5.
(a) A model representing the microenvironment from pH 4.5 to
7.3, where FAD is in the neutral oxidized form and His447 is in
the imidazole form protonated at NE2. (b) A model representing
the microenvironment at pH 9.0, where the negative charge of the
imidazolate form of His447 is stabilized by interaction with
Asn321 and Asn323.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nat Chem Biol
(2006,
2,
259-264)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Xin,
H.Yang,
X.Xia,
L.Zhang,
C.Cheng,
G.Mou,
J.Shi,
Y.Han,
and
W.Wang
(2011).
Affinity purification of a cholesterol oxidase expressed in Escherichia coli.
|
| |
J Chromatogr B Analyt Technol Biomed Life Sci,
879,
853-858.
|
 |
|
|
|
|
 |
P.Ferreira,
A.Hernandez-Ortega,
B.Herguedas,
A.T.Martínez,
and
M.Medina
(2009).
Aryl-alcohol oxidase involved in lignin degradation: a mechanistic study based on steady and pre-steady state kinetics and primary and solvent isotope effects with two alcohol substrates.
|
| |
J Biol Chem,
284,
24840-24847.
|
 |
|
|
|
|
 |
S.R.Thomas,
P.M.McTamney,
J.M.Adler,
N.Laronde-Leblanc,
and
S.E.Rokita
(2009).
Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands.
|
| |
J Biol Chem,
284,
19659-19667.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Chen,
A.Y.Lyubimov,
L.Brammer,
A.Vrielink,
and
N.S.Sampson
(2008).
The binding and release of oxygen and hydrogen peroxide are directed by a hydrophobic tunnel in cholesterol oxidase.
|
| |
Biochemistry,
47,
5368-5377.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Shemella,
B.Pereira,
Y.Zhang,
P.Van Roey,
G.Belfort,
S.Garde,
and
S.K.Nayak
(2007).
Mechanism for intein C-terminal cleavage: a proposal from quantum mechanical calculations.
|
| |
Biophys J,
92,
847-853.
|
 |
|
|
|
|
 |
L.De Colibus,
and
A.Mattevi
(2006).
New frontiers in structural flavoenzymology.
|
| |
Curr Opin Struct Biol,
16,
722-728.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

