 |
PDBsum entry 1mok

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1mok
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
NADPH dependent 2-ketopropyl coenzyme m oxidoreductase/carboxylase
|
|
Structure:
|
 |
Orf3. Chain: a, b, c, d. Synonym: 2-ketopropyl coenzyme m oxidoreductase/carboxylase. Ec: 1.8.1.5
|
|
Source:
|
 |
Xanthobacter autotrophicus. Organism_taxid: 78245. Strain: py2
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.225
|
R-free:
|
0.277
|
|
|
Authors:
|
 |
B.Nocek,S.B.Jang,M.S.Jeong,D.D.Clark,S.A.Ensign,J.W.Peters
|
Key ref:

|
 |
B.Nocek
et al.
(2002).
Structural basis for CO2 fixation by a novel member of the disulfide oxidoreductase family of enzymes, 2-ketopropyl-coenzyme M oxidoreductase/carboxylase.
Biochemistry,
41,
12907-12913.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
09-Sep-02
|
Release date:
|
27-Nov-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q56839
(XECC_XANP2) -
2-oxopropyl-CoM reductase, carboxylating from Xanthobacter autotrophicus (strain ATCC BAA-1158 / Py2)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
523 a.a.
522 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.8.1.5
- 2-oxopropyl-CoM reductase (carboxylating).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Epoxide Carboxylation
|
 |
 |
 |
 |
 |
Reaction:
|
 |
coenzyme M + acetoacetate + NADP+ = 2-oxopropyl-coenzyme M + CO2 + NADPH
|
 |
 |
 |
 |
 |
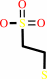 coenzyme M
coenzyme M
|
+
|
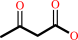 acetoacetate
acetoacetate
|
+
|
NADP(+)
Bound ligand (Het Group name = )
matches with 71.19% similarity
|
=
|
2-oxopropyl-coenzyme M
|
+
|
 CO2
CO2
|
+
|
 NADPH
NADPH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:12907-12913
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for CO2 fixation by a novel member of the disulfide oxidoreductase family of enzymes, 2-ketopropyl-coenzyme M oxidoreductase/carboxylase.
|
|
B.Nocek,
S.B.Jang,
M.S.Jeong,
D.D.Clark,
S.A.Ensign,
J.W.Peters.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The NADPH:2-ketopropyl-coenzyme M oxidoreductase/carboxylase (2-KPCC) is the
terminal enzyme in a metabolic pathway that results in the conversion of
propylene to the central metabolite acetoacetate in Xanthobacter autotrophicus
Py2. This enzyme is an FAD-containing enzyme that is a member of the
NADPH:disulfide oxidoreductase (DSOR) family of enzymes that include glutathione
reductase, dihydrolipoamide dehydrogenase, trypanothione reductase, thioredoxin
reductase, and mercuric reductase. In contrast to the prototypical reactions
catalyzed by members of the DSOR family, the NADPH:2-ketopropyl-coenzyme M
oxidoreductase/carboxylase catalyzes the reductive cleavage of the thioether
linkage of 2-ketopropyl-coenzyme M, and the subsequent carboxylation of the
ketopropyl cleavage product, yielding the products acetoacetate and free
coenzyme M. The structure of 2-KPCC reveals a unique active site in comparison
to those of other members of the DSOR family of enzymes and demonstrates how the
enzyme architecture has been adapted for the more sophisticated biochemical
reaction. In addition, comparison of the structures in the native state and in
the presence of bound substrate indicates the binding of the substrate
2-ketopropyl-coenzyme M induces a conformational change resulting in the
collapse of the substrate access channel. The encapsulation of the substrate in
this manner is reminiscent of the conformational changes observed in the
well-characterized CO2-fixing enzyme ribulose 1,5-bisphosphate
carboxylase/oxidase (Rubisco).

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.M.Krishnakumar,
D.Sliwa,
J.A.Endrizzi,
E.S.Boyd,
S.A.Ensign,
and
J.W.Peters
(2008).
Getting a handle on the role of coenzyme M in alkene metabolism.
|
| |
Microbiol Mol Biol Rev,
72,
445-456.
|
 |
|
|
|
|
 |
B.Nocek,
E.Evdokimova,
M.Proudfoot,
M.Kudritska,
L.L.Grochowski,
R.H.White,
A.Savchenko,
A.F.Yakunin,
A.Edwards,
and
A.Joachimiak
(2007).
Structure of an amide bond forming F(420):gamma-glutamyl ligase from Archaeoglobus fulgidus -- a member of a new family of non-ribosomal peptide synthases.
|
| |
J Mol Biol,
372,
456-469.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.M.Boyd,
A.Ellsworth,
and
S.A.Ensign
(2006).
Characterization of 2-bromoethanesulfonate as a selective inhibitor of the coenzyme m-dependent pathway and enzymes of bacterial aliphatic epoxide metabolism.
|
| |
J Bacteriol,
188,
8062-8069.
|
 |
|
|
|
|
 |
T.E.Mattes,
N.V.Coleman,
J.C.Spain,
and
J.M.Gossett
(2005).
Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614.
|
| |
Arch Microbiol,
183,
95.
|
 |
|
|
|
|
 |
S.A.Ensign,
and
J.R.Allen
(2003).
Aliphatic epoxide carboxylation.
|
| |
Annu Rev Biochem,
72,
55-76.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

