 |
PDBsum entry 1m5h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Formylmethanofuran:tetrahydromethanopterin formyltransferase from archaeoglobus fulgidus
|
|
Structure:
|
 |
Formylmethanofuran--tetrahydromethanopterin formyltransferase. Chain: a, b, c, d, e, f, g, h. Synonym: formylmethanofuran:tetrahydromethanopterin formyltransferase. Ftr-2. Ftr-2 af2207. Engineered: yes
|
|
Source:
|
 |
Archaeoglobus fulgidus. Organism_taxid: 2234. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Tetramer (from
)
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.229
|
R-free:
|
0.282
|
|
|
Authors:
|
 |
B.Mamat,A.Roth,C.Grimm,U.Ermler,C.Tziatzios,D.Schubert,R.K.Thauer, S.Shima
|
Key ref:

|
 |
B.Mamat
et al.
(2002).
Crystal structures and enzymatic properties of three formyltransferases from archaea: environmental adaptation and evolutionary relationship.
Protein Sci,
11,
2168-2178.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
09-Jul-02
|
Release date:
|
26-Jul-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O28076
(FTR_ARCFU) -
Formylmethanofuran--tetrahydromethanopterin formyltransferase from Archaeoglobus fulgidus (strain ATCC 49558 / DSM 4304 / JCM 9628 / NBRC 100126 / VC-16)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
297 a.a.
297 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.101
- formylmethanofuran--tetrahydromethanopterin N-formyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Methane Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-formylmethanofuran + 5,6,7,8-tetrahydromethanopterin + H+ = N5- formyl-5,6,7,8-tetrahydromethanopterin + methanofuran
|
 |
 |
 |
 |
 |
N-formylmethanofuran
|
+
|
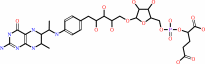 5,6,7,8-tetrahydromethanopterin
5,6,7,8-tetrahydromethanopterin
|
+
|
H(+)
|
=
|
N(5)- formyl-5,6,7,8-tetrahydromethanopterin
|
+
|
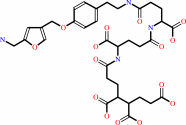 methanofuran
methanofuran
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
11:2168-2178
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures and enzymatic properties of three formyltransferases from archaea: environmental adaptation and evolutionary relationship.
|
|
B.Mamat,
A.Roth,
C.Grimm,
U.Ermler,
C.Tziatzios,
D.Schubert,
R.K.Thauer,
S.Shima.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Formyltransferase catalyzes the reversible formation of formylmethanofuran from
N(5)-formyltetrahydromethanopterin and methanofuran, a reaction involved in the
C1 metabolism of methanogenic and sulfate-reducing archaea. The crystal
structure of the homotetrameric enzyme from Methanopyrus kandleri (growth
temperature optimum 98 degrees C) has recently been solved at 1.65 A resolution.
We report here the crystal structures of the formyltransferase from
Methanosarcina barkeri (growth temperature optimum 37 degrees C) and from
Archaeoglobus fulgidus (growth temperature optimum 83 degrees C) at 1.9 A and
2.0 A resolution, respectively. Comparison of the structures of the three
enzymes revealed very similar folds. The most striking difference found was the
negative surface charge, which was -32 for the M. kandleri enzyme, only -8 for
the M. barkeri enzyme, and -11 for the A. fulgidus enzyme. The hydrophobic
surface fraction was 50% for the M. kandleri enzyme, 56% for the M. barkeri
enzyme, and 57% for the A. fulgidus enzyme. These differences most likely
reflect the adaptation of the enzyme to different cytoplasmic concentrations of
potassium cyclic 2,3-diphosphoglycerate, which are very high in M. kandleri (>1
M) and relatively low in M. barkeri and A. fulgidus. Formyltransferase is in a
monomer/dimer/tetramer equilibrium that is dependent on the salt concentration.
Only the dimers and tetramers are active, and only the tetramers are
thermostable. The enzyme from M. kandleri is a tetramer, which is active and
thermostable only at high concentrations of potassium phosphate (>1 M) or
potassium cyclic 2,3-diphosphoglycerate. Conversely, the enzyme from M. barkeri
and A. fulgidus already showed these properties, activity and stability, at much
lower concentrations of these strong salting-out salts.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Fig. 5. Structure of the formyltransferase. (A) The
tetramer presented as a Ribbon diagram indicates a particularly
extended contact region between subunits 1 (red) and 2 (green)
and the equivalent subunits 3 (blue) and 4 (orange). (B) The
Ribbon diagram of the monomer visualizes the location of the
insertion region (blue), the meander region (black circle), and
the loop between strands 6 and 7 (black arrow). (C) The stereo
C[  ]-plot of the
superimposed monomers of the enzymes from M. barkeri (red), A.
fulgidus (yellow), and M. kandleri (green) documents their
similar fold, in particular, in the core regions of the two
lobes. This figure was generated using the program MOLSCRIPT
(Kraulis 1991). ]-plot of the
superimposed monomers of the enzymes from M. barkeri (red), A.
fulgidus (yellow), and M. kandleri (green) documents their
similar fold, in particular, in the core regions of the two
lobes. This figure was generated using the program MOLSCRIPT
(Kraulis 1991).
|
 |
Figure 6.
Fig. 6. The electrostatic properties of the
formyltransferase tetramer from (A) M. barkeri, (B) A. fulgidus,
and (C) M. kandleri. The molecule surface is coated according to
the electrostatic potential: The extreme ranges of red and blue
represent potentials of -20k[B]T and 20k[B]T, respectively
(where k[B] is the Boltzmann constant and T is temperature). The
electrostatic surface potential of the enzymes from M. barkeri
and A. fulgidus is nearly neutral, and that of the M. kandleri
enzyme highly negative, reflecting the dominance of acidic to
basic residues. The potentials are calculated under salt-free
conditions. The figure was generated using the program GRASP
(Nicholls et al. 1993).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Protein Society:
Protein Sci
(2002,
11,
2168-2178)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.L.Powers,
C.R.Robinson,
and
A.S.Robinson
(2007).
Denaturation of an extremely stable hyperthermophilic protein occurs via a dimeric intermediate.
|
| |
Extremophiles,
11,
179-189.
|
 |
|
|
|
|
 |
R.Paulini,
K.Müller,
and
F.Diederich
(2005).
Orthogonal multipolar interactions in structural chemistry and biology.
|
| |
Angew Chem Int Ed Engl,
44,
1788-1805.
|
 |
|
|
|
|
 |
S.Sakasegawa,
C.H.Hagemeier,
R.K.Thauer,
L.O.Essen,
and
S.Shima
(2004).
Structural and functional analysis of the gpsA gene product of Archaeoglobus fulgidus: a glycerol-3-phosphate dehydrogenase with an unusual NADP+ preference.
|
| |
Protein Sci,
13,
3161-3171.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

