 |
PDBsum entry 1lvh

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.4.2.6
- beta-phosphoglucomutase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
b-phosphoglucomutase
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-glucose 1-phosphate = beta-D-glucose 6-phosphate
|
 |
 |
 |
 |
 |
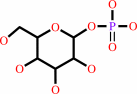 beta-D-glucose 1-phosphate
beta-D-glucose 1-phosphate
|
=
|
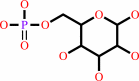 beta-D-glucose 6-phosphate
beta-D-glucose 6-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:8351-8359
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Caught in the act: the structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis.
|
|
S.D.Lahiri,
G.Zhang,
D.Dunaway-Mariano,
K.N.Allen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phosphoglucomutases catalyze the interconversion of D-glucose 1-phosphate and
D-glucose 6-phosphate, a reaction central to energy metabolism in all cells and
to the synthesis of cell wall polysaccharides in bacterial cells. Two classes of
phosphoglucomutases (alpha-PGM and beta-PGM) are distinguished on the basis of
their specificity for alpha- and beta-glucose-1-phosphate. beta-PGM is a member
of the haloacid dehalogenase (HAD) superfamily, which includes the sarcoplasmic
Ca(2+)-ATPase, phosphomannomutase, and phosphoserine phosphatase. beta-PGM is
unusual among family members in that the common phosphoenzyme intermediate
exists as a stable ground-state complex in this enzyme. Herein we report, for
the first time, the three-dimensional structure of a beta-PGM and the first view
of the true phosphoenzyme intermediate in the HAD superfamily. The crystal
structure of the Mg(II) complex of phosphorylated beta-phosphoglucomutase
(beta-PGM) from Lactococcus lactis has been determined to 2.3 A resolution by
multiwavelength anomalous diffraction (MAD) phasing on selenomethionine, and
refined to an R(cryst) = 0.24 and R(free) = 0.28. The active site of beta-PGM is
located between the core and the cap domain and is freely solvent accessible.
The residues within a 6 A radius of the phosphorylated Asp8 include Asp10,
Thr16, Ser114, Lys145, Glu169, and Asp170. The cofactor Mg(2+) is liganded with
octahedral coordination geometry by the carboxylate side chains of Asp8, Glu169,
Asp170, and the backbone carbonyl oxygen of Asp10 along with one oxygen from the
Asp8-phosphoryl group and one water ligand. The phosphate group of the
phosphoaspartyl residue, Asp8, interacts with the side chains of Ser114 and
Lys145. The absence of a base residue near the aspartyl phosphate group accounts
for the persistence of the phosphorylated enzyme under physiological conditions.
Substrate docking shows that glucose-6-P can bind to the active site of
phosphorylated beta-PGM in such a way as to position the C(1)OH near the
phosphoryl group of the phosphorylated Asp8 and the C(6) phosphoryl group near
the carboxylate group of Asp10. This result suggests a novel two-base mechanism
for phosphoryl group transfer in a phosphorylated sugar.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.V.Attwood,
P.G.Besant,
and
M.J.Piggott
(2011).
Focus on phosphoaspartate and phosphoglutamate.
|
| |
Amino Acids,
40,
1035-1051.
|
 |
|
|
|
|
 |
A.Preumont,
R.Rzem,
D.Vertommen,
and
E.Van Schaftingen
(2010).
HDHD1, which is often deleted in X-linked ichthyosis, encodes a pseudouridine-5'-phosphatase.
|
| |
Biochem J,
431,
237-244.
|
 |
|
|
|
|
 |
N.J.Baxter,
M.W.Bowler,
T.Alizadeh,
M.J.Cliff,
A.M.Hounslow,
B.Wu,
D.B.Berkowitz,
N.H.Williams,
G.M.Blackburn,
and
J.P.Waltho
(2010).
Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
|
| |
Proc Natl Acad Sci U S A,
107,
4555-4560.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Ye,
S.W.Crawley,
Y.Yang,
G.P.Côté,
and
Z.Jia
(2010).
Crystal structure of the alpha-kinase domain of Dictyostelium myosin heavy chain kinase A.
|
| |
Sci Signal,
3,
ra17.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Dai,
L.Finci,
C.Zhang,
S.Lahiri,
G.Zhang,
E.Peisach,
K.N.Allen,
and
D.Dunaway-Mariano
(2009).
Analysis of the structural determinants underlying discrimination between substrate and solvent in beta-phosphoglucomutase catalysis.
|
| |
Biochemistry,
48,
1984-1995.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.N.Allen,
and
D.Dunaway-Mariano
(2009).
Markers of fitness in a successful enzyme superfamily.
|
| |
Curr Opin Struct Biol,
19,
658-665.
|
 |
|
|
|
|
 |
T.Biswas,
L.Yi,
P.Aggarwal,
J.Wu,
J.R.Rubin,
J.A.Stuckey,
R.W.Woodard,
and
O.V.Tsodikov
(2009).
The tail of KdsC: conformational changes control the activity of a haloacid dehalogenase superfamily phosphatase.
|
| |
J Biol Chem,
284,
30594-30603.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.R.Diaz,
S.Stephenson,
J.M.Green,
V.M.Levdikov,
A.J.Wilkinson,
and
M.Perego
(2008).
Functional Role for a Conserved Aspartate in the Spo0E Signature Motif Involved in the Dephosphorylation of the Bacillus subtilis Sporulation Regulator Spo0A.
|
| |
J Biol Chem,
283,
2962-2972.
|
 |
|
|
|
|
 |
B.Soufi,
F.Gnad,
P.R.Jensen,
D.Petranovic,
M.Mann,
I.Mijakovic,
and
B.Macek
(2008).
The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins.
|
| |
Proteomics,
8,
3486-3493.
|
 |
|
|
|
|
 |
H.Yamamoto,
K.Takio,
M.Sugahara,
and
N.Kunishima
(2008).
Structure of a haloacid dehalogenase superfamily phosphatase PH1421 from Pyrococcus horikoshii OT3: oligomeric state and thermoadaptation mechanism.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
1068-1077.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.K.Bernstein,
F.Karimi-Busheri,
A.Rasouli-Nia,
R.Mani,
G.Dianov,
J.N.Glover,
and
M.Weinfeld
(2008).
Polynucleotide kinase as a potential target for enhancing cytotoxicity by ionizing radiation and topoisomerase I inhibitors.
|
| |
Anticancer Agents Med Chem,
8,
358-367.
|
 |
|
|
|
|
 |
R.Kluger,
and
S.Rathgeber
(2008).
Catalyzing separation of carbon dioxide in thiamin diphosphate-promoted decarboxylation.
|
| |
FEBS J,
275,
6089-6100.
|
 |
|
|
|
|
 |
Z.Lu,
D.Dunaway-Mariano,
and
K.N.Allen
(2008).
The catalytic scaffold of the haloalkanoic acid dehalogenase enzyme superfamily acts as a mold for the trigonal bipyramidal transition state.
|
| |
Proc Natl Acad Sci U S A,
105,
5687-5692.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Jemc,
and
I.Rebay
(2007).
The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription.
|
| |
Annu Rev Biochem,
76,
513-538.
|
 |
|
|
|
|
 |
L.Wang,
X.Yu,
P.Hu,
S.Broyde,
and
Y.Zhang
(2007).
A water-mediated and substrate-assisted catalytic mechanism for Sulfolobus solfataricus DNA polymerase IV.
|
| |
J Am Chem Soc,
129,
4731-4737.
|
 |
|
|
|
|
 |
A.R.Neves,
W.A.Pool,
R.Castro,
A.Mingote,
F.Santos,
J.Kok,
O.P.Kuipers,
and
H.Santos
(2006).
The alpha-phosphoglucomutase of Lactococcus lactis is unrelated to the alpha-D-phosphohexomutase superfamily and is encoded by the essential gene pgmH.
|
| |
J Biol Chem,
281,
36864-36873.
|
 |
|
|
|
|
 |
E.Kuznetsova,
M.Proudfoot,
C.F.Gonzalez,
G.Brown,
M.V.Omelchenko,
I.Borozan,
L.Carmel,
Y.I.Wolf,
H.Mori,
A.V.Savchenko,
C.H.Arrowsmith,
E.V.Koonin,
A.M.Edwards,
and
A.F.Yakunin
(2006).
Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family.
|
| |
J Biol Chem,
281,
36149-36161.
|
 |
|
|
|
|
 |
E.S.Groban,
A.Narayanan,
and
M.P.Jacobson
(2006).
Conformational changes in protein loops and helices induced by post-translational phosphorylation.
|
| |
PLoS Comput Biol,
2,
e32.
|
 |
|
|
|
|
 |
E.S.Rangarajan,
A.Proteau,
J.Wagner,
M.N.Hung,
A.Matte,
and
M.Cygler
(2006).
Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
|
| |
J Biol Chem,
281,
37930-37941.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.N.Rao,
D.Kumaran,
J.Seetharaman,
J.B.Bonanno,
S.K.Burley,
and
S.Swaminathan
(2006).
Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications.
|
| |
Protein Sci,
15,
1735-1744.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.J.Baxter,
L.F.Olguin,
M.Golicnik,
G.Feng,
A.M.Hounslow,
W.Bermel,
G.M.Blackburn,
F.Hollfelder,
J.P.Waltho,
and
N.H.Williams
(2006).
A Trojan horse transition state analogue generated by MgF3- formation in an enzyme active site.
|
| |
Proc Natl Acad Sci U S A,
103,
14732-14737.
|
 |
|
|
|
|
 |
N.R.Silvaggi,
C.Zhang,
Z.Lu,
J.Dai,
D.Dunaway-Mariano,
and
K.N.Allen
(2006).
The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
|
| |
J Biol Chem,
281,
14918-14926.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Lin,
L.C.Pedersen,
V.K.Batra,
W.A.Beard,
S.H.Wilson,
and
L.G.Pedersen
(2006).
Energy analysis of chemistry for correct insertion by DNA polymerase beta.
|
| |
Proc Natl Acad Sci U S A,
103,
13294-13299.
|
 |
|
|
|
|
 |
S.D.Lahiri,
G.Zhang,
D.Dunaway-Mariano,
and
K.N.Allen
(2006).
Diversification of function in the haloacid dehalogenase enzyme superfamily: The role of the cap domain in hydrolytic phosphoruscarbon bond cleavage.
|
| |
Bioorg Chem,
34,
394-409.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Roberts,
S.Y.Lee,
E.McCullagh,
R.E.Silversmith,
and
D.E.Wemmer
(2005).
YbiV from Escherichia coli K12 is a HAD phosphatase.
|
| |
Proteins,
58,
790-801.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Kuznetsova,
M.Proudfoot,
S.A.Sanders,
J.Reinking,
A.Savchenko,
C.H.Arrowsmith,
A.M.Edwards,
and
A.F.Yakunin
(2005).
Enzyme genomics: Application of general enzymatic screens to discover new enzymes.
|
| |
FEMS Microbiol Rev,
29,
263-279.
|
 |
|
|
|
|
 |
F.J.Sandoval,
and
S.Roje
(2005).
An FMN hydrolase is fused to a riboflavin kinase homolog in plants.
|
| |
J Biol Chem,
280,
38337-38345.
|
 |
|
|
|
|
 |
A.Barth,
and
N.Bezlyepkina
(2004).
P-O bond destabilization accelerates phosphoenzyme hydrolysis of sarcoplasmic reticulum Ca2+ -ATPase.
|
| |
J Biol Chem,
279,
51888-51896.
|
 |
|
|
|
|
 |
C.Toyoshima,
and
G.Inesi
(2004).
Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum.
|
| |
Annu Rev Biochem,
73,
269-292.
|
 |
|
|
|
|
 |
C.Toyoshima,
and
T.Mizutani
(2004).
Crystal structure of the calcium pump with a bound ATP analogue.
|
| |
Nature,
430,
529-535.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.C.Meng,
B.J.Polacco,
and
P.C.Babbitt
(2004).
Superfamily active site templates.
|
| |
Proteins,
55,
962-976.
|
 |
|
|
|
|
 |
P.H.Tartaix,
M.Doulaverakis,
A.George,
L.W.Fisher,
W.T.Butler,
C.Qin,
E.Salih,
M.Tan,
Y.Fujimoto,
L.Spevak,
and
A.L.Boskey
(2004).
In vitro effects of dentin matrix protein-1 on hydroxyapatite formation provide insights into in vivo functions.
|
| |
J Biol Chem,
279,
18115-18120.
|
 |
|
|
|
|
 |
S.K.Singh,
K.Yang,
S.Karthikeyan,
T.Huynh,
X.Zhang,
M.A.Phillips,
and
H.Zhang
(2004).
The thrH gene product of Pseudomonas aeruginosa is a dual activity enzyme with a novel phosphoserine:homoserine phosphotransferase activity.
|
| |
J Biol Chem,
279,
13166-13173.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Peeraer,
A.Rabijns,
J.F.Collet,
E.Van Schaftingen,
and
C.De Ranter
(2004).
How calcium inhibits the magnesium-dependent enzyme human phosphoserine phosphatase.
|
| |
Eur J Biochem,
271,
3421-3427.
|
 |
|
|
|
|
 |
D.H.Shin,
A.Roberts,
J.Jancarik,
H.Yokota,
R.Kim,
D.E.Wemmer,
and
S.H.Kim
(2003).
Crystal structure of a phosphatase with a unique substrate binding domain from Thermotoga maritima.
|
| |
Protein Sci,
12,
1464-1472.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Ma,
G.Inesi,
and
C.Toyoshima
(2003).
Substrate-induced conformational fit and headpiece closure in the Ca2+ATPase (SERCA).
|
| |
J Biol Chem,
278,
28938-28943.
|
 |
|
|
|
|
 |
S.D.Lahiri,
G.Zhang,
D.Dunaway-Mariano,
and
K.N.Allen
(2003).
The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction.
|
| |
Science,
299,
2067-2071.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

