
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of the adenylyl cyclase domain of anthrax edema factor (ef) in complex with calmodulin and 2' deoxy, 3' anthraniloyl atp
|
|
Structure:
|
 |
Calmodulin-sensitive adenylate cyclase. Chain: a, b, c. Fragment: c-terminal domain (residues 291-800). Synonym: atp pyrophosphate-lyase, adenylyl cyclase, edema factor, ef, anthrax edema toxin adenylate cyclase component. Engineered: yes. Calmodulin. Chain: d, e, f. Engineered: yes
|
|
Source:
|
 |
Bacillus anthracis. Organism_taxid: 1392. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693. Homo sapiens. Human. Organism_taxid: 9606.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
3.60Å
|
R-factor:
|
0.281
|
R-free:
|
0.307
|
|
|
Authors:
|
 |
Y.Shen,Y.-S.Lee,S.Soelaiman,P.Bergson,D.Lu,A.Chen,K.Beckingham, Z.Grabarek,M.Mrksich,W.-J.Tang
|
Key ref:

|
 |
Y.Shen
et al.
(2002).
Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins.
EMBO J,
21,
6721-6732.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
28-May-02
|
Release date:
|
04-Dec-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C:
E.C.4.6.1.1
- adenylate cyclase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
ATP = 3',5'-cyclic AMP + diphosphate
|
 |
 |
 |
 |
 |
ATP
Bound ligand (Het Group name = )
matches with 75.00% similarity
|
=
|
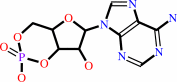 3',5'-cyclic AMP
3',5'-cyclic AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
21:6721-6732
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins.
|
|
Y.Shen,
Y.S.Lee,
S.Soelaiman,
P.Bergson,
D.Lu,
A.Chen,
K.Beckingham,
Z.Grabarek,
M.Mrksich,
W.J.Tang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Edema factor (EF) and CyaA are calmodulin (CaM)-activated adenylyl cyclase
exotoxins involved in the pathogenesis of anthrax and whooping cough,
respectively. Using spectroscopic, enzyme kinetic and surface plasmon resonance
spectroscopy analyses, we show that low Ca(2+) concentrations increase the
affinity of CaM for EF and CyaA causing their activation, but higher Ca(2+)
concentrations directly inhibit catalysis. Both events occur in a
physiologically relevant range of Ca(2+) concentrations. Despite the similarity
in Ca(2+) sensitivity, EF and CyaA have substantial differences in CaM binding
and activation. CyaA has 100-fold higher affinity for CaM than EF. CaM has N-
and C-terminal globular domains, each binding two Ca(2+) ions. CyaA can be fully
activated by CaM mutants with one defective C-terminal Ca(2+)-binding site or by
either terminal domain of CaM while EF cannot. EF consists of a catalytic core
and a helical domain, and both are required for CaM activation of EF. Mutations
that decrease the interaction of the helical domain with the catalytic core
create an enzyme with higher sensitivity to Ca(2+)-CaM activation. However, CyaA
is fully activated by CaM without the domain corresponding to the helical domain
of EF.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4 The activation of EF and CyaA-N by wild-type CaM and
two series of CaM mutants. EF (1 nM) and CyaA-N (0.7 nM) were
used for an adenylyl cyclase activity assay in the presence of
0.1  M
free Ca^2+ with CaM mutants CaM 41/75 and 85/112. Each mutant
has two cysteine mutations to lock either the N- or the
C-terminal domain of CaM in the closed conformation (A and B).
The same concentrations of EF and CyaA-N were used for an
adenylyl cyclase activity assay in the presence of 1 M
free Ca^2+ with CaM mutants CaM 41/75 and 85/112. Each mutant
has two cysteine mutations to lock either the N- or the
C-terminal domain of CaM in the closed conformation (A and B).
The same concentrations of EF and CyaA-N were used for an
adenylyl cyclase activity assay in the presence of 1  M
free Ca^2+ with CaM mutants B1Q, B2Q, B3Q and B4Q. Each mutant
has a mutation inactivating one of four calcium-binding sites (C
and D). Means M
free Ca^2+ with CaM mutants B1Q, B2Q, B3Q and B4Q. Each mutant
has a mutation inactivating one of four calcium-binding sites (C
and D). Means  SE
are representative of at least two experiments. SE
are representative of at least two experiments.
|
 |
Figure 5.
Figure 5 Effect of calcium and magnesium ions on adenylyl
cyclase activity of EF and CyaA-N (A and B) and an EF mutant,
EF-H577N (C and D). Adenylyl cyclase activity assays were
performed with 10  M
CaM at 0.1 M
CaM at 0.1  M
Ca^2+ (filled circles), 0.3 M
Ca^2+ (filled circles), 0.3  M
Ca^2+ (open circles) and 1.0 M
Ca^2+ (open circles) and 1.0  M
Ca^2+ (filled triangles) in the presence of 1 nM EF (A), and 0.7
nM CyaA-N (B). To analyze the mutant form of EF, adenylyl
cyclase activities were measured with 10 M
Ca^2+ (filled triangles) in the presence of 1 nM EF (A), and 0.7
nM CyaA-N (B). To analyze the mutant form of EF, adenylyl
cyclase activities were measured with 10  M
CaM and 1 nM EF or 66 nM EF-H577N with either 0.3 M
CaM and 1 nM EF or 66 nM EF-H577N with either 0.3  M
Ca^2+ (C) or 10 mM Mg^2+ (D). Both were buffered by 10 mM EGTA.
Maximal activities for EF in the magnesium titration are 1616
s^-1 (0.1 M
Ca^2+ (C) or 10 mM Mg^2+ (D). Both were buffered by 10 mM EGTA.
Maximal activities for EF in the magnesium titration are 1616
s^-1 (0.1  M
Ca^2+), 1074 s^-1 (0.3 M
Ca^2+), 1074 s^-1 (0.3  M
Ca^2+), and 1009 s^-1 (1.0 M
Ca^2+), and 1009 s^-1 (1.0  M
Ca^2+) (A) and those for CyaA-N are 2106 s^-1 (0.1 M
Ca^2+) (A) and those for CyaA-N are 2106 s^-1 (0.1  M
Ca^2+), 1674 s^-1 (0.3 M
Ca^2+), 1674 s^-1 (0.3  M
Ca^2+) and 1385 s^-1 (1.0 M
Ca^2+) and 1385 s^-1 (1.0  M)
(B). Maximal activities for EF and EF-H577N were 1208 s^-1 and 4
s^-1 (C) and those for EF and EF-H577N were 2726 s^-1 and 6 s^-1
(D), respectively. Means M)
(B). Maximal activities for EF and EF-H577N were 1208 s^-1 and 4
s^-1 (C) and those for EF and EF-H577N were 2726 s^-1 and 6 s^-1
(D), respectively. Means  SE
are representative of at least two experiments. SE
are representative of at least two experiments.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(2002,
21,
6721-6732)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Martínez,
T.E.Malliavin,
and
A.Blondel
(2011).
Mechanism of reactant and product dissociation from the anthrax edema factor: A locally enhanced sampling and steered molecular dynamics study.
|
| |
Proteins,
79,
1649-1661.
|
 |
|
|
|
|
 |
E.Laine,
C.Goncalves,
J.C.Karst,
A.Lesnard,
S.Rault,
W.J.Tang,
T.E.Malliavin,
D.Ladant,
and
A.Blondel
(2010).
Use of allostery to identify inhibitors of calmodulin-induced activation of Bacillus anthracis edema factor.
|
| |
Proc Natl Acad Sci U S A,
107,
11277-11282.
|
 |
|
|
|
|
 |
H.M.Taha,
J.Schmidt,
M.Göttle,
S.Suryanarayana,
Y.Shen,
W.J.Tang,
A.Gille,
J.Geduhn,
B.König,
S.Dove,
and
R.Seifert
(2009).
Molecular analysis of the interaction of anthrax adenylyl cyclase toxin, edema factor, with 2'(3')-O-(N-(methyl)anthraniloyl)-substituted purine and pyrimidine nucleotides.
|
| |
Mol Pharmacol,
75,
693-703.
|
 |
|
|
|
|
 |
L.Martínez,
E.Laine,
T.E.Malliavin,
M.Nilges,
and
A.Blondel
(2009).
ATP conformations and ion binding modes in the active site of anthrax edema factor: a computational analysis.
|
| |
Proteins,
77,
971-983.
|
 |
|
|
|
|
 |
S.Suryanarayana,
J.L.Wang,
M.Richter,
Y.Shen,
W.J.Tang,
G.H.Lushington,
and
R.Seifert
(2009).
Distinct interactions of 2'- and 3'-O-(N-methyl)anthraniloyl-isomers of ATP and GTP with the adenylyl cyclase toxin of Bacillus anthracis, edema factor.
|
| |
Biochem Pharmacol,
78,
224-230.
|
 |
|
|
|
|
 |
W.J.Tang,
and
Q.Guo
(2009).
The adenylyl cyclase activity of anthrax edema factor.
|
| |
Mol Aspects Med,
30,
423-430.
|
 |
|
|
|
|
 |
Z.Chen,
M.Moayeri,
H.Zhao,
D.Crown,
S.H.Leppla,
and
R.H.Purcell
(2009).
Potent neutralization of anthrax edema toxin by a humanized monoclonal antibody that competes with calmodulin for edema factor binding.
|
| |
Proc Natl Acad Sci U S A,
106,
13487-13492.
|
 |
|
|
|
|
 |
J.Gsponer,
J.Christodoulou,
A.Cavalli,
J.M.Bui,
B.Richter,
C.M.Dobson,
and
M.Vendruscolo
(2008).
A coupled equilibrium shift mechanism in calmodulin-mediated signal transduction.
|
| |
Structure,
16,
736-746.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Guo,
J.E.Jureller,
J.T.Warren,
E.Solomaha,
J.Florián,
and
W.J.Tang
(2008).
Protein-protein docking and analysis reveal that two homologous bacterial adenylyl cyclase toxins interact with calmodulin differently.
|
| |
J Biol Chem,
283,
23836-23845.
|
 |
|
|
|
|
 |
J.T.Warren,
Q.Guo,
and
W.J.Tang
(2007).
A 1.3-A structure of zinc-bound N-terminal domain of calmodulin elucidates potential early ion-binding step.
|
| |
J Mol Biol,
374,
517-527.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Fiser,
J.Masín,
M.Basler,
J.Krusek,
V.Spuláková,
I.Konopásek,
and
P.Sebo
(2007).
Third activity of Bordetella adenylate cyclase (AC) toxin-hemolysin. Membrane translocation of AC domain polypeptide promotes calcium influx into CD11b+ monocytes independently of the catalytic and hemolytic activities.
|
| |
J Biol Chem,
282,
2808-2820.
|
 |
|
|
|
|
 |
J.Vojtova,
J.Kamanova,
and
P.Sebo
(2006).
Bordetella adenylate cyclase toxin: a swift saboteur of host defense.
|
| |
Curr Opin Microbiol,
9,
69-75.
|
 |
|
|
|
|
 |
K.Chen,
J.Ruan,
and
L.A.Kurgan
(2006).
Prediction of three dimensional structure of calmodulin.
|
| |
Protein J,
25,
57-70.
|
 |
|
|
|
|
 |
Q.Guo,
Y.Shen,
Y.S.Lee,
C.S.Gibbs,
M.Mrksich,
and
W.J.Tang
(2005).
Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin.
|
| |
EMBO J,
24,
3190-3201.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Shen,
N.L.Zhukovskaya,
Q.Guo,
J.Florián,
and
W.J.Tang
(2005).
Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
|
| |
EMBO J,
24,
929-941.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Gille,
G.H.Lushington,
T.C.Mou,
M.B.Doughty,
R.A.Johnson,
and
R.Seifert
(2004).
Differential inhibition of adenylyl cyclase isoforms and soluble guanylyl cyclase by purine and pyrimidine nucleotides.
|
| |
J Biol Chem,
279,
19955-19969.
|
 |
|
|
|
|
 |
H.Barth,
K.Aktories,
M.R.Popoff,
and
B.G.Stiles
(2004).
Binary bacterial toxins: biochemistry, biology, and applications of common Clostridium and Bacillus proteins.
|
| |
Microbiol Mol Biol Rev,
68,
373.
|
 |
|
|
|
|
 |
Q.Guo,
Y.Shen,
N.L.Zhukovskaya,
J.Florián,
and
W.J.Tang
(2004).
Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate.
|
| |
J Biol Chem,
279,
29427-29435.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Vougier,
J.Mary,
N.Dautin,
J.Vinh,
B.Friguet,
and
D.Ladant
(2004).
Essential role of methionine residues in calmodulin binding to Bordetella pertussis adenylate cyclase, as probed by selective oxidation and repair by the peptide methionine sulfoxide reductases.
|
| |
J Biol Chem,
279,
30210-30218.
|
 |
|
|
|
|
 |
Y.S.Lee,
P.Bergson,
W.S.He,
M.Mrksich,
and
W.J.Tang
(2004).
Discovery of a small molecule that inhibits the interaction of anthrax edema factor with its cellular activator, calmodulin.
|
| |
Chem Biol,
11,
1139-1146.
|
 |
|
|
|
|
 |
Y.Shen,
N.L.Zhukovskaya,
M.I.Zimmer,
S.Soelaiman,
P.Bergson,
C.R.Wang,
C.S.Gibbs,
and
W.J.Tang
(2004).
Selective inhibition of anthrax edema factor by adefovir, a drug for chronic hepatitis B virus infection.
|
| |
Proc Natl Acad Sci U S A,
101,
3242-3247.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.J.Collier,
and
J.A.Young
(2003).
Anthrax toxin.
|
| |
Annu Rev Cell Dev Biol,
19,
45-70.
|
 |
|
|
|
|
 |
R.L.Rich,
and
D.G.Myszka
(2003).
A survey of the year 2002 commercial optical biosensor literature.
|
| |
J Mol Recognit,
16,
351-382.
|
 |
|
|
|
|
 |
S.Soelaiman,
B.Q.Wei,
P.Bergson,
Y.S.Lee,
Y.Shen,
M.Mrksich,
B.K.Shoichet,
and
W.J.Tang
(2003).
Structure-based inhibitor discovery against adenylyl cyclase toxins from pathogenic bacteria that cause anthrax and whooping cough.
|
| |
J Biol Chem,
278,
25990-25997.
|
 |
|
|
|
|
 |
T.S.Ulmer,
S.Soelaiman,
S.Li,
C.B.Klee,
W.J.Tang,
and
A.Bax
(2003).
Calcium dependence of the interaction between calmodulin and anthrax edema factor.
|
| |
J Biol Chem,
278,
29261-29266.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

