 |
PDBsum entry 1lpp

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.3
- triacylglycerol lipase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a triacylglycerol + H2O = a diacylglycerol + a fatty acid + H+
|
 |
 |
 |
 |
 |
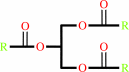 triacylglycerol
triacylglycerol
|
+
|
H2O
|
=
|
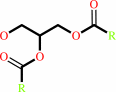 diacylglycerol
diacylglycerol
|
+
|
 fatty acid
fatty acid
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
33:3494-3500
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Analogs of reaction intermediates identify a unique substrate binding site in Candida rugosa lipase.
|
|
P.Grochulski,
F.Bouthillier,
R.J.Kazlauskas,
A.N.Serreqi,
J.D.Schrag,
E.Ziomek,
M.Cygler.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structures of Candida rugosa lipase-inhibitor complexes demonstrate that the
scissile fatty acyl chain is bound in a narrow, hydrophobic tunnel which is
unique among lipases studied to date. Modeling of triglyceride binding suggests
that the bound lipid must adopt a "tuning fork" conformation. The complexes,
analogs of tetrahedral intermediates of the acylation and deacylation steps of
the reaction pathway, localize the components of the oxyanion hole and define
the stereochemistry of ester hydrolysis. Comparison with other lipases suggests
that the positioning of the scissile fatty acyl chain and ester bond and the
stereochemistry of hydrolysis are the same in all lipases which share the
alpha/beta-hydrolase fold.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Jiang,
K.L.Morley,
J.D.Schrag,
and
R.J.Kazlauskas
(2011).
Different active-site loop orientation in serine hydrolases versus acyltransferases.
|
| |
Chembiochem,
12,
768-776.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Bakolitsa,
A.Kumar,
D.McMullan,
S.S.Krishna,
M.D.Miller,
D.Carlton,
R.Najmanovich,
P.Abdubek,
T.Astakhova,
H.J.Chiu,
T.Clayton,
M.C.Deller,
L.Duan,
Y.Elias,
J.Feuerhelm,
J.C.Grant,
S.K.Grzechnik,
G.W.Han,
L.Jaroszewski,
K.K.Jin,
H.E.Klock,
M.W.Knuth,
P.Kozbial,
D.Marciano,
A.T.Morse,
E.Nigoghossian,
L.Okach,
S.Oommachen,
J.Paulsen,
R.Reyes,
C.L.Rife,
C.V.Trout,
H.van den Bedem,
D.Weekes,
A.White,
Q.Xu,
K.O.Hodgson,
J.Wooley,
M.A.Elsliger,
A.M.Deacon,
A.Godzik,
S.A.Lesley,
and
I.A.Wilson
(2010).
The structure of the first representative of Pfam family PF06475 reveals a new fold with possible involvement in glycolipid metabolism.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
66,
1211-1217.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Simón,
F.M.Muñiz,
A.F.de Arriba,
V.Alcázar,
C.Raposo,
and
J.R.Morán
(2010).
Synthesis of a chiral artificial receptor with catalytic activity in Michael additions and its chiral resolution by a new methodology.
|
| |
Org Biomol Chem,
8,
1763-1768.
|
 |
|
|
|
|
 |
M.D.Benaiges,
M.Alarcón,
P.Fuciños,
P.Ferrer,
M.Rua,
and
F.Valero
(2010).
Recombinant Candida rugosa lipase 2 from Pichia pastoris: immobilization and use as biocatalyst in a stereoselective reaction.
|
| |
Biotechnol Prog,
26,
1252-1258.
|
 |
|
|
|
|
 |
P.B.Juhl,
P.Trodler,
S.Tyagi,
and
J.Pleiss
(2009).
Modelling substrate specificity and enantioselectivity for lipases and esterases by substrate-imprinted docking.
|
| |
BMC Struct Biol,
9,
39.
|
 |
|
|
|
|
 |
A.Maeda,
T.Mizuno,
M.Bunya,
S.Sugihara,
D.Nakayama,
S.Tsunasawa,
Y.Hirota,
and
A.Sugihara
(2008).
Characterization of novel cholesterol esterase from Trichoderma sp. AS59 with high ability to synthesize steryl esters.
|
| |
J Biosci Bioeng,
105,
341-349.
|
 |
|
|
|
|
 |
L.Mandrich,
V.Menchise,
V.Alterio,
G.De Simone,
C.Pedone,
M.Rossi,
and
G.Manco
(2008).
Functional and structural features of the oxyanion hole in a thermophilic esterase from Alicyclobacillus acidocaldarius.
|
| |
Proteins,
71,
1721-1731.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Kasrayan,
M.Bocola,
A.G.Sandström,
G.Lavén,
and
J.E.Bäckvall
(2007).
Prediction of the Candida antarctica lipase A protein structure by comparative modeling and site-directed mutagenesis.
|
| |
Chembiochem,
8,
1409-1415.
|
 |
|
|
|
|
 |
B.R.Somashekar,
K.Lohith,
B.Manohar,
and
S.Divakar
(2007).
Inhibition of Rhizomucor miehei and Candida rugosa lipases by D-glucose in esterification between L-alanine and D-glucose.
|
| |
J Biosci Bioeng,
103,
122-128.
|
 |
|
|
|
|
 |
G.Lin,
W.C.Liao,
and
Z.H.Ku
(2005).
Quantitative structure-activity relationships for the pre-steady state of Pseudomonas species lipase inhibitions by p-nirophenyl-N-substituted carbamates.
|
| |
Protein J,
24,
201-207.
|
 |
|
|
|
|
 |
J.Aishima,
D.S.Russel,
L.J.Guibas,
P.D.Adams,
and
A.T.Brunger
(2005).
Automated crystallographic ligand building using the medial axis transform of an electron-density isosurface.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1354-1363.
|
 |
|
|
|
|
 |
S.Y.Chiou,
C.Y.Lai,
L.Y.Lin,
and
G.Lin
(2005).
Probing stereoselective inhibition of the acyl binding site of cholesterol esterase with four diastereomers of 2'-N-alpha-methylbenzylcarbamyl-1, 1'-bi-2-naphthol.
|
| |
BMC Biochem,
6,
17.
|
 |
|
|
|
|
 |
C.C.Akoh,
G.C.Lee,
and
J.F.Shaw
(2004).
Protein engineering and applications of Candida rugosa lipase isoforms.
|
| |
Lipids,
39,
513-526.
|
 |
|
|
|
|
 |
C.C.Akoh,
G.C.Lee,
Y.C.Liaw,
T.H.Huang,
and
J.F.Shaw
(2004).
GDSL family of serine esterases/lipases.
|
| |
Prog Lipid Res,
43,
534-552.
|
 |
|
|
|
|
 |
G.De Simone,
L.Mandrich,
V.Menchise,
V.Giordano,
F.Febbraio,
M.Rossi,
C.Pedone,
and
G.Manco
(2004).
A substrate-induced switch in the reaction mechanism of a thermophilic esterase: kinetic evidences and structural basis.
|
| |
J Biol Chem,
279,
6815-6823.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.López,
M.A.Pernas,
L.M.Pastrana,
A.Sánchez,
F.Valero,
and
M.L.Rúa
(2004).
Reactivity of pure Candida rugosa lipase isoenzymes (Lip1, Lip2, and Lip3) in aqueous and organic media. influence of the isoenzymatic profile on the lipase performance in organic media.
|
| |
Biotechnol Prog,
20,
65-73.
|
 |
|
|
|
|
 |
A.Kobayashi,
Y.Sato,
and
F.Mizutani
(2001).
Adsorption properties and activities of lipase on a gold substrate modified by self-assembled monolayers.
|
| |
Biosci Biotechnol Biochem,
65,
2392-2396.
|
 |
|
|
|
|
 |
P.Berglund
(2001).
Controlling lipase enantioselectivity for organic synthesis.
|
| |
Biomol Eng,
18,
13-22.
|
 |
|
|
|
|
 |
T.Osterlund
(2001).
Structure-function relationships of hormone-sensitive lipase.
|
| |
Eur J Biochem,
268,
1899-1907.
|
 |
|
|
|
|
 |
A.Svendsen
(2000).
Lipase protein engineering.
|
| |
Biochim Biophys Acta,
1543,
223-238.
|
 |
|
|
|
|
 |
C.N.Lin,
and
S.W.Tsai
(2000).
Dynamic kinetic resolution of suprofen thioester via coupled trioctylamine and lipase catalysis.
|
| |
Biotechnol Bioeng,
69,
31-38.
|
 |
|
|
|
|
 |
G.Lin,
W.C.Liao,
and
S.Y.Chiou
(2000).
Quantitative structure-activity relationships for the pre-steady-state inhibition of cholesterol esterase by 4-nitrophenyl-N-substituted carbamates.
|
| |
Bioorg Med Chem,
8,
2601-2607.
|
 |
|
|
|
|
 |
S.Brocca,
M.Persson,
E.Wehtje,
P.Adlercreutz,
L.Alberghina,
and
M.Lotti
(2000).
Mutants provide evidence of the importance of glycosydic chains in the activation of lipase 1 from Candida rugosa.
|
| |
Protein Sci,
9,
985-990.
|
 |
|
|
|
|
 |
S.Terzyan,
C.S.Wang,
D.Downs,
B.Hunter,
and
X.C.Zhang
(2000).
Crystal structure of the catalytic domain of human bile salt activated lipase.
|
| |
Protein Sci,
9,
1783-1790.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Z.Pletnev,
T.S.Zamolodchikova,
W.A.Pangborn,
and
W.L.Duax
(2000).
Crystal structure of bovine duodenase, a serine protease, with dual trypsin and chymotrypsin-like specificities.
|
| |
Proteins,
41,
8.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Cajal,
A.Svendsen,
J.De Bolós,
S.A.Patkar,
and
M.A.Alsina
(2000).
Effect of the lipid interface on the catalytic activity and spectroscopic properties of a fungal lipase.
|
| |
Biochimie,
82,
1053-1061.
|
 |
|
|
|
|
 |
Y.Cajal,
A.Svendsen,
V.Girona,
S.A.Patkar,
and
M.A.Alsina
(2000).
Interfacial control of lid opening in Thermomyces lanuginosa lipase.
|
| |
Biochemistry,
39,
413-423.
|
 |
|
|
|
|
 |
A.Roussel,
S.Canaan,
M.P.Egloff,
M.Rivière,
L.Dupuis,
R.Verger,
and
C.Cambillau
(1999).
Crystal structure of human gastric lipase and model of lysosomal acid lipase, two lipolytic enzymes of medical interest.
|
| |
J Biol Chem,
274,
16995-17002.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Lin,
C.T.Shieh,
Y.C.Tsai,
C.I.Hwang,
C.P.Lu,
and
G.H.Chen
(1999).
Structure-reactivity probes for active site shapes of cholesterol esterase by carbamate inhibitors.
|
| |
Biochim Biophys Acta,
1431,
500-511.
|
 |
|
|
|
|
 |
G.Zandonella,
P.Stadler,
L.Haalck,
F.Spener,
F.Paltauf,
and
A.Hermetter
(1999).
Interactions of fluorescent triacylglycerol analogs covalently bound to the active site of a lipase from Rhizopus oryzae.
|
| |
Eur J Biochem,
262,
63-69.
|
 |
|
|
|
|
 |
J.W.Simons,
R.C.Cox,
M.R.Egmond,
and
H.M.Verheij
(1999).
Rational design of alpha-keto triglyceride analogues as inhibitors for Staphylococcus hyicus lipase.
|
| |
Biochemistry,
38,
6346-6351.
|
 |
|
|
|
|
 |
K.E.Jaeger,
B.W.Dijkstra,
and
M.T.Reetz
(1999).
Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases.
|
| |
Annu Rev Microbiol,
53,
315-351.
|
 |
|
|
|
|
 |
A.Ordentlich,
D.Barak,
C.Kronman,
N.Ariel,
Y.Segall,
B.Velan,
and
A.Shafferman
(1998).
Functional characteristics of the oxyanion hole in human acetylcholinesterase.
|
| |
J Biol Chem,
273,
19509-19517.
|
 |
|
|
|
|
 |
D.Mileto,
S.Brocca,
M.Lotti,
M.Takagi,
C.Alquati,
and
L.Alberghina
(1998).
Characterization of the Candida rugosa lipase system and overexpression of the lip1 isoenzyme in a non-conventional yeast.
|
| |
Chem Phys Lipids,
93,
47-55.
|
 |
|
|
|
|
 |
F.Haeffner,
T.Norin,
and
K.Hult
(1998).
Molecular modeling of the enantioselectivity in lipase-catalyzed transesterification reactions.
|
| |
Biophys J,
74,
1251-1262.
|
 |
|
|
|
|
 |
G.Lin,
Y.C.Tsai,
H.C.Liu,
W.C.Liao,
and
C.H.Chang
(1998).
Enantiomeric inhibitors of cholesterol esterase and acetylcholinesterase.
|
| |
Biochim Biophys Acta,
1388,
161-174.
|
 |
|
|
|
|
 |
H.Hirohara,
and
M.Nishizawa
(1998).
Biochemical synthesis of several chiral insecticide intermediates and mechanisms of action of relevant enzymes.
|
| |
Biosci Biotechnol Biochem,
62,
1-9.
|
 |
|
|
|
|
 |
J.Pleiss,
M.Fischer,
and
R.D.Schmid
(1998).
Anatomy of lipase binding sites: the scissile fatty acid binding site.
|
| |
Chem Phys Lipids,
93,
67-80.
|
 |
|
|
|
|
 |
M.Holmquist
(1998).
Insights into the molecular basis for fatty acyl specificities of lipases from Geotrichum candidum and Candida rugosa.
|
| |
Chem Phys Lipids,
93,
57-66.
|
 |
|
|
|
|
 |
Z.Keresztessy,
and
M.A.Hughes
(1998).
Homology modelling and molecular dynamics aided analysis of ligand complexes demonstrates functional properties of lipid-transfer proteins encoded by the barley low-temperature-inducible gene family, blt4.
|
| |
Plant J,
14,
523-533.
|
 |
|
|
|
|
 |
J.D.Schrag,
Y.Li,
M.Cygler,
D.Lang,
T.Burgdorf,
H.J.Hecht,
R.Schmid,
D.Schomburg,
T.J.Rydel,
J.D.Oliver,
L.C.Strickland,
C.M.Dunaway,
S.B.Larson,
J.Day,
and
A.McPherson
(1997).
The open conformation of a Pseudomonas lipase.
|
| |
Structure,
5,
187-202.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.K.Kim,
H.K.Song,
D.H.Shin,
K.Y.Hwang,
and
S.W.Suh
(1997).
The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor.
|
| |
Structure,
5,
173-185.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.R.Feaster,
D.M.Quinn,
and
B.L.Barnett
(1997).
Molecular modeling of the structures of human and rat pancreatic cholesterol esterases.
|
| |
Protein Sci,
6,
73-79.
|
 |
|
|
|
|
 |
A.Nicolas,
M.Egmond,
C.T.Verrips,
J.de Vlieg,
S.Longhi,
C.Cambillau,
and
C.Martinez
(1996).
Contribution of cutinase serine 42 side chain to the stabilization of the oxyanion transition state.
|
| |
Biochemistry,
35,
398-410.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Holmquist,
F.Haeffner,
T.Norin,
and
K.Hult
(1996).
A structural basis for enantioselective inhibition of Candida rugosa lipase by long-chain aliphatic alcohols.
|
| |
Protein Sci,
5,
83-88.
|
 |
|
|
|
|
 |
S.Jääskeläinen,
X.Y.Wu,
S.Linko,
Y.Wang,
Y.Y.Linko,
O.Teleman,
and
P.Linko
(1996).
Production, characterization, and molecular modeling of lipases for esterification.
|
| |
Ann N Y Acad Sci,
799,
129-138.
|
 |
|
|
|
|
 |
D.Ghosh,
Z.Wawrzak,
V.Z.Pletnev,
N.Li,
R.Kaiser,
W.Pangborn,
H.Jörnvall,
M.Erman,
and
W.L.Duax
(1995).
Structure of uncomplexed and linoleate-bound Candida cylindracea cholesterol esterase.
|
| |
Structure,
3,
279-288.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.H.Shin,
J.Y.Lee,
K.Y.Hwang,
K.K.Kim,
and
S.W.Suh
(1995).
High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings.
|
| |
Structure,
3,
189-199.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.C.Bertolini,
J.D.Schrag,
M.Cygler,
E.Ziomek,
D.Y.Thomas,
and
T.Vernet
(1995).
Expression and characterization of Geotrichum candidum lipase I gene. Comparison of specificity profile with lipase II.
|
| |
Eur J Biochem,
228,
863-869.
|
 |
|
|
|
|
 |
M.Cygler,
P.Grochulski,
and
J.D.Schrag
(1995).
Structural determinants defining common stereoselectivity of lipases toward secondary alcohols.
|
| |
Can J Microbiol,
41,
289-296.
|
 |
|
|
|
|
 |
M.Norin,
F.Haeffner,
A.Achour,
T.Norin,
and
K.Hult
(1994).
Computer modeling of substrate binding to lipases from Rhizomucor miehei, Humicola lanuginosa, and Candida rugosa.
|
| |
Protein Sci,
3,
1493-1503.
|
 |
|
|
|
|
 |
R.J.Kazlauskas
(1994).
Elucidating structure-mechanism relationships in lipases: prospects for predicting and engineering catalytic properties.
|
| |
Trends Biotechnol,
12,
464-472.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

