 |
PDBsum entry 1kv3

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Human tissue transglutaminase in gdp bound form
|
|
Structure:
|
 |
Protein-glutamine gamma-glutamyltransferase. Chain: a, b, c, d, e, f. Synonym: tissue transglutaminase, tgasE C, tgc, tgase-h. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: tgm2. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.233
|
R-free:
|
0.272
|
|
|
Authors:
|
 |
S.Liu,R.A.Cerione,J.Clardy
|
Key ref:

|
 |
S.Liu
et al.
(2002).
Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity.
Proc Natl Acad Sci U S A,
99,
2743-2747.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
24-Jan-02
|
Release date:
|
13-Mar-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P21980
(TGM2_HUMAN) -
Protein-glutamine gamma-glutamyltransferase 2 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
687 a.a.
651 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.3.2.13
- protein-glutamine gamma-glutamyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutaminyl-[protein] + L-lysyl-[protein] = [protein]-L-lysyl-N6-5- L-glutamyl-[protein] + NH4+
|
 |
 |
 |
 |
 |
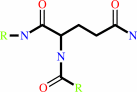 protein-L-glutamine
protein-L-glutamine
|
+
|
protein-L-lysine
|
=
|
protein with an N(6)- (gamma-glutamyl)-L-lysine cross-link
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Ca(2+)
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.5.1.44
- protein-glutamine glutaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutaminyl-[protein] + H2O = L-glutamyl-[protein] + NH4+
|
 |
 |
 |
 |
 |
 Protein L-glutamine
Protein L-glutamine
|
+
|
H(2)O
|
=
|
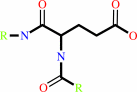 protein L-glutamate
protein L-glutamate
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
99:2743-2747
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity.
|
|
S.Liu,
R.A.Cerione,
J.Clardy.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Tissue transglutaminase (TG) is a Ca2+-dependent acyltransferase with roles in
cellular differentiation, apoptosis, and other biological functions. In addition
to being a transamidase, TG undergoes a GTP-binding/GTPase cycle even though it
lacks any obvious sequence similarity with canonical GTP-binding (G) proteins.
Guanine nucleotide binding and Ca2+ concentration reciprocally regulate TG's
transamidation activity, with nucleotide binding being the negative regulator.
Here we report the x-ray structure determined to 2.8-A resolution of human TG
complexed with GDP. Although the transamidation active site is similar to those
of other known transglutaminases, the guanine nucleotide-binding site of TG
differs markedly from other G proteins. The structure suggests a structural
basis for the negative regulation of transamidation activity by bound
nucleotide, and the positive regulation of transamidation by Ca2+.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Comparisons between the atomic interactions of
GDP with TG (Left) and Ras (Right). Hydrogen bonds and ion pair
interactions are shown in dashed lines. The GDP molecule is
shown in ball-and-stick. TG and Ras residues are shown in thin
sticks. Drawing prepared with MOLSCRIPT (44) and RASTER3D (45).
|
 |
Figure 6.
Fig. 6. Comparison of the calcium-binding sites of TG
(green) and factor XIIIa (red). In factor XIIIa, the loop
involved in calcium binding is oriented toward the Ca^2+-binding
site, whereas in TG-GDP, the same loop is oriented toward GDP.
Figure prepared with MOLSCRIPT (44) and RASTER3D (45).
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Png,
G.K.Samivelu,
S.H.Yeo,
J.Chew,
S.S.Chaurasia,
and
L.Tong
(2011).
Hyperosmolarity-mediated mitochondrial dysfunction requires Transglutaminase-2 in human corneal epithelial cells.
|
| |
J Cell Physiol,
226,
693-699.
|
 |
|
|
|
|
 |
G.Colak,
J.W.Keillor,
and
G.V.Johnson
(2011).
Cytosolic Guanine nucledotide binding deficient form of transglutaminase 2 (r580a) potentiates cell death in oxygen glucose deprivation.
|
| |
PLoS One,
6,
e16665.
|
 |
|
|
|
|
 |
I.Komáromi,
Z.Bagoly,
and
L.Muszbek
(2011).
Factor XIII: novel structural and functional aspects.
|
| |
J Thromb Haemost,
9,
9.
|
 |
|
|
|
|
 |
L.Dafik,
and
C.Khosla
(2011).
Dihydroisoxazole analogs for labeling and visualization of catalytically active transglutaminase 2.
|
| |
Chem Biol,
18,
58-66.
|
 |
|
|
|
|
 |
A.E.Tee,
G.M.Marshall,
P.Y.Liu,
N.Xu,
M.Haber,
M.D.Norris,
S.E.Iismaa,
and
T.Liu
(2010).
Opposing effects of two tissue transglutaminase protein isoforms in neuroblastoma cell differentiation.
|
| |
J Biol Chem,
285,
3561-3567.
|
 |
|
|
|
|
 |
C.M.Bergamini,
A.Dondi,
V.Lanzara,
M.Squerzanti,
C.Cervellati,
K.Montin,
C.Mischiati,
G.Tasco,
R.Collighan,
M.Griffin,
and
R.Casadio
(2010).
Thermodynamics of binding of regulatory ligands to tissue transglutaminase.
|
| |
Amino Acids,
39,
297-304.
|
 |
|
|
|
|
 |
D.Park,
S.S.Choi,
and
K.S.Ha
(2010).
Transglutaminase 2: a multi-functional protein in multiple subcellular compartments.
|
| |
Amino Acids,
39,
619-631.
|
 |
|
|
|
|
 |
H.Tatsukawa,
and
S.Kojima
(2010).
Recent advances in understanding the roles of transglutaminase 2 in alcoholic steatohepatitis.
|
| |
Cell Biol Int,
34,
325-334.
|
 |
|
|
|
|
 |
S.P.Bustos,
and
R.A.Reithmeier
(2010).
Protein 4.2 interaction with hereditary spherocytosis mutants of the cytoplasmic domain of human anion exchanger 1.
|
| |
Biochem J,
433,
313-322.
|
 |
|
|
|
|
 |
T.S.Lai,
C.Davies,
and
C.S.Greenberg
(2010).
Human tissue transglutaminase is inhibited by pharmacologic and chemical acetylation.
|
| |
Protein Sci,
19,
229-235.
|
 |
|
|
|
|
 |
A.Scarpellini,
R.Germack,
H.Lortat-Jacob,
T.Muramatsu,
E.Billett,
T.Johnson,
and
E.A.Verderio
(2009).
Heparan sulfate proteoglycans are receptors for the cell-surface trafficking and biological activity of transglutaminase-2.
|
| |
J Biol Chem,
284,
18411-18423.
|
 |
|
|
|
|
 |
D.A.Bachovchin,
S.J.Brown,
H.Rosen,
and
B.F.Cravatt
(2009).
Identification of selective inhibitors of uncharacterized enzymes by high-throughput screening with fluorescent activity-based probes.
|
| |
Nat Biotechnol,
27,
387-394.
|
 |
|
|
|
|
 |
E.A.Zemskov,
E.Loukinova,
I.Mikhailenko,
R.A.Coleman,
D.K.Strickland,
and
A.M.Belkin
(2009).
Regulation of platelet-derived growth factor receptor function by integrin-associated cell surface transglutaminase.
|
| |
J Biol Chem,
284,
16693-16703.
|
 |
|
|
|
|
 |
R.J.Collighan,
and
M.Griffin
(2009).
Transglutaminase 2 cross-linking of matrix proteins: biological significance and medical applications.
|
| |
Amino Acids,
36,
659-670.
|
 |
|
|
|
|
 |
S.Gundemir,
and
G.V.Johnson
(2009).
Intracellular localization and conformational state of transglutaminase 2: implications for cell death.
|
| |
PLoS One,
4,
e6123.
|
 |
|
|
|
|
 |
S.J.Yi,
J.Groffen,
and
N.Heisterkamp
(2009).
Transglutaminase 2 regulates the GTPase-activating activity of Bcr.
|
| |
J Biol Chem,
284,
35645-35651.
|
 |
|
|
|
|
 |
T.M.Jeitner,
N.A.Muma,
K.P.Battaile,
and
A.J.Cooper
(2009).
Transglutaminase activation in neurodegenerative diseases.
|
| |
Future Neurol,
4,
449-467.
|
 |
|
|
|
|
 |
D.S.Wang,
D.W.Dickson,
and
J.S.Malter
(2008).
Tissue Transglutaminase, Protein Cross-linking and Alzheimer's Disease: Review and Views.
|
| |
Int J Clin Exp Pathol,
1,
5.
|
 |
|
|
|
|
 |
Q.Ruan,
J.Tucholski,
S.Gundemir,
and
G.V.Johnson Voll
(2008).
The Differential Effects of R580A Mutation on Transamidation and GTP Binding Activity of Rat and Human Type 2 Transglutaminase.
|
| |
Int J Clin Exp Med,
1,
248-259.
|
 |
|
|
|
|
 |
T.S.Lai,
Y.Liu,
T.Tucker,
K.R.Daniel,
D.C.Sane,
E.Toone,
J.R.Burke,
W.J.Strittmatter,
and
C.S.Greenberg
(2008).
Identification of chemical inhibitors to human tissue transglutaminase by screening existing drug libraries.
|
| |
Chem Biol,
15,
969-978.
|
 |
|
|
|
|
 |
V.Pietroni,
S.Di Giorgi,
A.Paradisi,
B.Ahvazi,
E.Candi,
and
G.Melino
(2008).
Inactive and highly active, proteolytically processed transglutaminase-5 in epithelial cells.
|
| |
J Invest Dermatol,
128,
2760-2766.
|
 |
|
|
|
|
 |
D.M.Pinkas,
P.Strop,
A.T.Brunger,
and
C.Khosla
(2007).
Transglutaminase 2 undergoes a large conformational change upon activation.
|
| |
PLoS Biol,
5,
e327.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Langston,
A.Blinkovsky,
T.Byun,
M.Terribilini,
D.Ransbarger,
and
F.Xu
(2007).
Substrate specificity of streptomyces transglutaminases.
|
| |
Appl Biochem Biotechnol,
136,
291-308.
|
 |
|
|
|
|
 |
K.M.Boeshans,
T.C.Mueser,
and
B.Ahvazi
(2007).
A three-dimensional model of the human transglutaminase 1: insights into the understanding of lamellar ichthyosis.
|
| |
J Mol Model,
13,
233-246.
|
 |
|
|
|
|
 |
M.Siegel,
and
C.Khosla
(2007).
Transglutaminase 2 inhibitors and their therapeutic role in disease states.
|
| |
Pharmacol Ther,
115,
232-245.
|
 |
|
|
|
|
 |
S.Datta,
M.A.Antonyak,
and
R.A.Cerione
(2007).
GTP-binding-defective forms of tissue transglutaminase trigger cell death.
|
| |
Biochemistry,
46,
14819-14829.
|
 |
|
|
|
|
 |
T.S.Lai,
Y.Liu,
W.Li,
and
C.S.Greenberg
(2007).
Identification of two GTP-independent alternatively spliced forms of tissue transglutaminase in human leukocytes, vascular smooth muscle, and endothelial cells.
|
| |
FASEB J,
21,
4131-4143.
|
 |
|
|
|
|
 |
A.Janiak,
E.A.Zemskov,
and
A.M.Belkin
(2006).
Cell surface transglutaminase promotes RhoA activation via integrin clustering and suppression of the Src-p190RhoGAP signaling pathway.
|
| |
Mol Biol Cell,
17,
1606-1619.
|
 |
|
|
|
|
 |
D.M.Rose,
A.D.Sydlaske,
A.Agha-Babakhani,
K.Johnson,
and
R.Terkeltaub
(2006).
Transglutaminase 2 limits murine peritoneal acute gout-like inflammation by regulating macrophage clearance of apoptotic neutrophils.
|
| |
Arthritis Rheum,
54,
3363-3371.
|
 |
|
|
|
|
 |
G.E.Begg,
L.Carrington,
P.H.Stokes,
J.M.Matthews,
M.A.Wouters,
A.Husain,
L.Lorand,
S.E.Iismaa,
and
R.M.Graham
(2006).
Mechanism of allosteric regulation of transglutaminase 2 by GTP.
|
| |
Proc Natl Acad Sci U S A,
103,
19683-19688.
|
 |
|
|
|
|
 |
J.J.Wakshlag,
M.A.Antonyak,
J.E.Boehm,
K.Boehm,
and
R.A.Cerione
(2006).
Effects of tissue transglutaminase on beta -amyloid1-42-induced apoptosis.
|
| |
Protein J,
25,
83-94.
|
 |
|
|
|
|
 |
S.Datta,
M.A.Antonyak,
and
R.A.Cerione
(2006).
Importance of Ca(2+)-dependent transamidation activity in the protection afforded by tissue transglutaminase against doxorubicin-induced apoptosis.
|
| |
Biochemistry,
45,
13163-13174.
|
 |
|
|
|
|
 |
Z.Keresztessy,
E.Csosz,
J.Hársfalvi,
K.Csomós,
J.Gray,
R.N.Lightowlers,
J.H.Lakey,
Z.Balajthy,
and
L.Fésüs
(2006).
Phage display selection of efficient glutamine-donor substrate peptides for transglutaminase 2.
|
| |
Protein Sci,
15,
2466-2480.
|
 |
|
|
|
|
 |
C.Esposito,
and
I.Caputo
(2005).
Mammalian transglutaminases. Identification of substrates as a key to physiological function and physiopathological relevance.
|
| |
FEBS J,
272,
615-631.
|
 |
|
|
|
|
 |
C.D.Bailey,
and
G.V.Johnson
(2004).
Developmental regulation of tissue transglutaminase in the mouse forebrain.
|
| |
J Neurochem,
91,
1369-1379.
|
 |
|
|
|
|
 |
K.T.Barglow,
and
B.F.Cravatt
(2004).
Discovering disease-associated enzymes by proteome reactivity profiling.
|
| |
Chem Biol,
11,
1523-1531.
|
 |
|
|
|
|
 |
R.A.Chica,
P.Gagnon,
J.W.Keillor,
and
J.N.Pelletier
(2004).
Tissue transglutaminase acylation: Proposed role of conserved active site Tyr and Trp residues revealed by molecular modeling of peptide substrate binding.
|
| |
Protein Sci,
13,
979-991.
|
 |
|
|
|
|
 |
J.H.Jeon,
K.H.Choi,
S.Y.Cho,
C.W.Kim,
D.M.Shin,
J.C.Kwon,
K.Y.Song,
S.C.Park,
and
I.G.Kim
(2003).
Transglutaminase 2 inhibits Rb binding of human papillomavirus E7 by incorporating polyamine.
|
| |
EMBO J,
22,
5273-5282.
|
 |
|
|
|
|
 |
L.Lorand,
and
R.M.Graham
(2003).
Transglutaminases: crosslinking enzymes with pleiotropic functions.
|
| |
Nat Rev Mol Cell Biol,
4,
140-156.
|
 |
|
|
|
|
 |
S.E.Iismaa,
S.Holman,
M.A.Wouters,
L.Lorand,
R.M.Graham,
and
A.Husain
(2003).
Evolutionary specialization of a tryptophan indole group for transition-state stabilization by eukaryotic transglutaminases.
|
| |
Proc Natl Acad Sci U S A,
100,
12636-12641.
|
 |
|
|
|
|
 |
D.Sblattero,
F.Florian,
E.Azzoni,
T.Zyla,
M.Park,
V.Baldas,
T.Not,
A.Ventura,
A.Bradbury,
and
R.Marzari
(2002).
The analysis of the fine specificity of celiac disease antibodies using tissue transglutaminase fragments.
|
| |
Eur J Biochem,
269,
5175-5181.
|
 |
|
|
|
|
 |
F.Brunner,
S.Rosahl,
J.Lee,
J.J.Rudd,
C.Geiler,
S.Kauppinen,
G.Rasmussen,
D.Scheel,
and
T.Nürnberger
(2002).
Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases.
|
| |
EMBO J,
21,
6681-6688.
|
 |
|
|
|
|
 |
F.Wada,
A.Nakamura,
T.Masutani,
K.Ikura,
M.Maki,
and
K.Hitomi
(2002).
Identification of mammalian-type transglutaminase in Physarum polycephalum. Evidence from the cDNA sequence and involvement of GTP in the regulation of transamidating activity.
|
| |
Eur J Biochem,
269,
3451-3460.
|
 |
|
|
|
|
 |
L.Fesus,
and
M.Piacentini
(2002).
Transglutaminase 2: an enigmatic enzyme with diverse functions.
|
| |
Trends Biochem Sci,
27,
534-539.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

