 |
PDBsum entry 1kmt

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Protein binding
|
PDB id
|
|

|

|
1kmt
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Protein binding
|
 |
|
Title:
|
 |
Crystal structure of rhogdi glu(154,155)ala mutant
|
|
Structure:
|
 |
Rho gdp-dissociation inhibitor 1. Chain: a, b. Synonym: rho gdi 1, rho-gdi alpha. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.30Å
|
R-factor:
|
0.160
|
R-free:
|
0.195
|
|
|
Authors:
|
 |
A.Mateja,Y.Devedjiev,D.Krowarsh,K.Longenecker,Z.Dauter,J.Otlewski, Z.S.Derewenda
|
Key ref:

|
 |
A.Mateja
et al.
(2002).
The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution.
Acta Crystallogr D Biol Crystallogr,
58,
1983-1991.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
17-Dec-01
|
Release date:
|
11-Dec-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P52565
(GDIR1_HUMAN) -
Rho GDP-dissociation inhibitor 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
204 a.a.
138 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 4 residue positions (black
crosses)
|
|

|
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
58:1983-1991
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution.
|
|
A.Mateja,
Y.Devedjiev,
D.Krowarsch,
K.Longenecker,
Z.Dauter,
J.Otlewski,
Z.S.Derewenda.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
It is hypothesized that surface residues with high conformational entropy,
specifically lysines and glutamates, impede protein crystallization. In a
previous study using a model system of Rho-specific guanine nucleotide
dissociation inhibitor (RhoGDI), it was shown that mutating Lys residues to Ala
results in enhanced crystallizability, particularly when clusters of lysines are
targeted. It was also shown that one of these mutants formed crystals that
yielded diffraction to 2.0 A, a significant improvement on the wild-type protein
crystals. In the current paper, an analysis of the impact of surface mutations
replacing Glu residues with Ala or Asp on the stability and crystallization
properties of RhoGDI is presented. The Glu-->Ala (Asp) mutants are generally
more likely to produce crystals of the protein than the wild-type and in one
case the resulting crystals yielded a diffraction pattern to 1.2 A resolution.
This occurs in spite of the fact that mutating surface Glu residues almost
invariably affects the protein's stability, as illustrated by the reduced deltaG
between folded and unfolded forms measured by isothermal equilibrium
denaturation. The present study strongly supports the notion that rational
surface mutagenesis can be an effective tool in overcoming problems stemming
from the protein's recalcitrance to crystallization and may also yield dramatic
improvements in crystal quality.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 A view of the RhoGDI 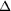 66
fragment with glutamates shown in full and colored gold. The two
sites which were mutated in the highly diffracting crystal form
are shown in pink. The sites are numbered according to the
amino-acid sequence of the full-length protein. The figure was
prepared using RIBBONS (Carson, 1991[Carson, M. (1991). J. Appl.
Cryst. 24, 958-961.]). 66
fragment with glutamates shown in full and colored gold. The two
sites which were mutated in the highly diffracting crystal form
are shown in pink. The sites are numbered according to the
amino-acid sequence of the full-length protein. The figure was
prepared using RIBBONS (Carson, 1991[Carson, M. (1991). J. Appl.
Cryst. 24, 958-961.]).
|
 |
Figure 3.
Figure 3 Ribbon diagram of the overall arrangement of the
molecules in the asymmetric unit, with the location of the point
mutations identified by magenta spheres. The two molecules
within the asymmetric unit are colored according to the
temperature factors. The grey sphere indicates the entrance to
the hydrophobic pocket. The figure was prepared using BOBSCRIPT
(Esnouf, 1997[Esnouf, R. M. (1997). J. Mol. Graph. 15,
132-143.]).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2002,
58,
1983-1991)
copyright 2002.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Schalk-Hihi,
C.Schubert,
R.Alexander,
S.Bayoumy,
J.C.Clemente,
I.Deckman,
R.L.DesJarlais,
K.C.Dzordzorme,
C.M.Flores,
B.Grasberger,
J.K.Kranz,
F.Lewandowski,
L.Liu,
H.Ma,
D.Maguire,
M.J.Macielag,
M.E.McDonnell,
T.Mezzasalma Haarlander,
R.Miller,
C.Milligan,
C.Reynolds,
and
L.C.Kuo
(2011).
Crystal structure of a soluble form of human monoglyceride lipase in complex with an inhibitor at 1.35 Å resolution.
|
| |
Protein Sci,
20,
670-683.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.S.Derewenda
(2011).
It's all in the crystals….
|
| |
Acta Crystallogr D Biol Crystallogr,
67,
243-248.
|
 |
|
|
|
|
 |
Z.S.Derewenda
(2010).
Application of protein engineering to enhance crystallizability and improve crystal properties.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
604-615.
|
 |
|
|
|
|
 |
M.Alexander,
M.Gerauer,
M.Pechlivanis,
B.Popkirova,
R.Dvorsky,
L.Brunsveld,
H.Waldmann,
and
J.Kuhlmann
(2009).
Mapping the isoprenoid binding pocket of PDEdelta by a semisynthetic, photoactivatable N-Ras lipoprotein.
|
| |
Chembiochem,
10,
98.
|
 |
|
|
|
|
 |
S.Das,
R.M.Yennamalli,
A.Vishnoi,
P.Gupta,
and
A.Bhattacharya
(2009).
Single-nucleotide variations associated with Mycobacterium tuberculosis KwaZulu-Natal strains.
|
| |
J Biosci,
34,
397-404.
|
 |
|
|
|
|
 |
T.Shimamura,
Y.Nitanai,
T.Uchiyama,
and
H.Matsuzawa
(2009).
Improvement of crystal quality by surface mutations of beta-lactamase Toho-1.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
379-382.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.N.Price,
Y.Chen,
S.K.Handelman,
H.Neely,
P.Manor,
R.Karlin,
R.Nair,
J.Liu,
M.Baran,
J.Everett,
S.N.Tong,
F.Forouhar,
S.S.Swaminathan,
T.Acton,
R.Xiao,
J.R.Luft,
A.Lauricella,
G.T.DeTitta,
B.Rost,
G.T.Montelione,
and
J.F.Hunt
(2009).
Understanding the physical properties that control protein crystallization by analysis of large-scale experimental data.
|
| |
Nat Biotechnol,
27,
51-57.
|
 |
|
|
|
|
 |
B.A.Manjasetty,
A.P.Turnbull,
S.Panjikar,
K.Büssow,
and
M.R.Chance
(2008).
Automated technologies and novel techniques to accelerate protein crystallography for structural genomics.
|
| |
Proteomics,
8,
612-625.
|
 |
|
|
|
|
 |
M.Senda,
S.Muto,
M.Horikoshi,
and
T.Senda
(2008).
Effect of leucine-to-methionine substitutions on the diffraction quality of histone chaperone SET/TAF-Ibeta/INHAT crystals.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
960-965.
|
 |
|
|
|
|
 |
B.Liu,
V.M.Luna,
Y.Chen,
C.D.Stout,
and
J.A.Fee
(2007).
An unexpected outcome of surface engineering an integral membrane protein: improved crystallization of cytochrome ba(3) from Thermus thermophilus.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
1029-1034.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Shaw,
C.Cheng,
W.Tempel,
J.Chang,
J.Ng,
X.Y.Wang,
S.Perrett,
J.Rose,
Z.Rao,
B.C.Wang,
and
Z.J.Liu
(2007).
(NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
|
| |
BMC Struct Biol,
7,
46.
|
 |
|
|
|
|
 |
G.Roos,
E.Brosens,
K.Wahni,
A.Desmyter,
S.Spinelli,
L.Wyns,
J.Messens,
and
R.Loris
(2006).
Combining site-specific mutagenesis and seeding as a strategy to crystallize 'difficult' proteins: the case of Staphylococcus aureus thioredoxin.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
1255-1258.
|
 |
|
|
|
|
 |
T.S.Walter,
C.Meier,
R.Assenberg,
K.F.Au,
J.Ren,
A.Verma,
J.E.Nettleship,
R.J.Owens,
D.I.Stuart,
and
J.M.Grimes
(2006).
Lysine methylation as a routine rescue strategy for protein crystallization.
|
| |
Structure,
14,
1617-1622.
|
 |
|
|
|
|
 |
D.M.Anstrom,
L.Colip,
B.Moshofsky,
E.Hatcher,
and
S.J.Remington
(2005).
Systematic replacement of lysine with glutamine and alanine in Escherichia coli malate synthase G: effect on crystallization.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
1069-1074.
|
 |
|
|
|
|
 |
E.Dransart,
B.Olofsson,
and
J.Cherfils
(2005).
RhoGDIs revisited: novel roles in Rho regulation.
|
| |
Traffic,
6,
957-966.
|
 |
|
|
|
|
 |
R.J.Keenan,
D.L.Siehl,
R.Gorton,
and
L.A.Castle
(2005).
DNA shuffling as a tool for protein crystallization.
|
| |
Proc Natl Acad Sci U S A,
102,
8887-8892.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.P.Doye,
A.A.Louis,
and
M.Vendruscolo
(2004).
Inhibition of protein crystallization by evolutionary negative design.
|
| |
Phys Biol,
1,
P9-13.
|
 |
|
|
|
|
 |
T.U.Schwartz,
R.Walczak,
and
G.Blobel
(2004).
Circular permutation as a tool to reduce surface entropy triggers crystallization of the signal recognition particle receptor beta subunit.
|
| |
Protein Sci,
13,
2814-2818.
|
 |
|
|
|
|
 |
U.Derewenda,
A.Mateja,
Y.Devedjiev,
K.M.Routzahn,
A.G.Evdokimov,
Z.S.Derewenda,
and
D.S.Waugh
(2004).
The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague.
|
| |
Structure,
12,
301-306.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.S.Derewenda
(2004).
Rational protein crystallization by mutational surface engineering.
|
| |
Structure,
12,
529-535.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

