 |
PDBsum entry 1jue

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1jue
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.98.1
- dihydroorotate oxidase (fumarate).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-dihydroorotate + fumarate = orotate + succinate
|
 |
 |
 |
 |
 |
 (S)-dihydroorotate
(S)-dihydroorotate
|
+
|
fumarate
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
=
|
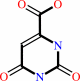 orotate
orotate
|
+
|
 succinate
succinate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:28812-28822
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function.
|
|
S.Nørager,
S.Arent,
O.Björnberg,
M.Ottosen,
L.Lo Leggio,
K.F.Jensen,
S.Larsen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Dihydroorotate dehydrogenases (DHODs) are flavoenzymes catalyzing the oxidation
of (S)-dihydroorotate to orotate in the biosynthesis of UMP, the precursor of
all other pyrimidine nucleotides. On the basis of sequence, DHODs can be divided
into two classes, class 1, further divided in subclasses 1A and 1B, and class 2.
This division corresponds to differences in cellular location and the nature of
the electron acceptor. Herein we report a study of Lactococcus lactis DHODA, a
representative of the class 1A enzymes. Based on the DHODA structure we selected
seven residues that are highly conserved between both main classes of DHODs as
well as three residues representing surface charges close to the active site for
site-directed mutagenesis. The availability of both kinetic and structural data
on the mutant enzymes allowed us to define the roles individual structural
segments play in catalysis. We have also structurally proven the presence of an
open active site loop in DHODA and obtained information about the interactions
that control movements of loops around the active site. Furthermore, in one
mutant structure we observed differences between the two monomers of the dimer,
confirming an apparent asymmetry between the two substrate binding sites that
was indicated by the kinetic results.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIG. 1. Structures of the native and variant DHODAs. Top,
the native DHODA dimer and the residues chosen for mutations.
FMN (yellow) and orotate (orange) are shown as stick models. The
N- and C-terminals of the two subunits of the dimer are
indicated with an N and C, respectively. The catalytic active
base Cys-130 and the mutated residues are illustrated as stick
models and are color-coded according to their location in the
sequence. Blue: Arg-50, Pro-56, Arg-57, and the cis-proline loop
(42-58); pink: Ser-129, Cys-130, Pro-131, Lys-136, and the
active site loop (129-138); violet: Asn-127 and the  -strand
123-127; green: Asn-67 and the loop 67-75; turquoise: Asn-193
and the loop 191-195; and red: Lys-213 and the Lys-213-helix
(211-214). Bottom, the A subunit of the native structure in the
presence of DTT and absence of orotate and three mutant
structures (K213E(Oro), P56A(Oro) and K136E) oriented as the
dimer at the top and color-coded according to the temperature
factors of the residues. The color code goes from blue to red
with blue representing residues with B-factors below or equal to
5 Å2 and red corresponding to B-factors above or equal to
55 Å2. -strand
123-127; green: Asn-67 and the loop 67-75; turquoise: Asn-193
and the loop 191-195; and red: Lys-213 and the Lys-213-helix
(211-214). Bottom, the A subunit of the native structure in the
presence of DTT and absence of orotate and three mutant
structures (K213E(Oro), P56A(Oro) and K136E) oriented as the
dimer at the top and color-coded according to the temperature
factors of the residues. The color code goes from blue to red
with blue representing residues with B-factors below or equal to
5 Å2 and red corresponding to B-factors above or equal to
55 Å2.
|
 |
Figure 4.
FIG. 4. Selected close-up views of native and mutant
DHODAs. a, alignment of the subunit A of native DHODA orotate
complex (violet) and the K213E(Oro) structure (green). FMN and
orotate are shown as yellow and orange stick models,
respectively. The figure visualizes the difference between the
open active site loop and the closed active site loop. A
different view of the active site highlighting the interactions
between protein and orotate (in orange) is shown for the native
orotate complex (b) and N67A(Oro) (c), with the FMN group
colored in magenta. Hydrogen bonds are shown as black dotted
lines and selected water molecules are represented as cyan
spheres.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
28812-28822)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Nisimoto,
H.M.Jackson,
H.Ogawa,
T.Kawahara,
and
J.D.Lambeth
(2010).
Constitutive NADPH-dependent electron transferase activity of the Nox4 dehydrogenase domain.
|
| |
Biochemistry,
49,
2433-2442.
|
 |
|
|
|
|
 |
R.L.Fagan,
and
B.A.Palfey
(2009).
Roles in binding and chemistry for conserved active site residues in the class 2 dihydroorotate dehydrogenase from Escherichia coli.
|
| |
Biochemistry,
48,
7169-7178.
|
 |
|
|
|
|
 |
R.L.Kow,
J.R.Whicher,
C.A.McDonald,
B.A.Palfey,
and
R.L.Fagan
(2009).
Disruption of the proton relay network in the class 2 dihydroorotate dehydrogenase from Escherichia coli.
|
| |
Biochemistry,
48,
9801-9809.
|
 |
|
|
|
|
 |
W.Y.Liu,
M.M.Wang,
J.Huang,
H.J.Tang,
H.X.Lan,
and
H.S.Zhang
(2009).
The OsDHODH1 gene is involved in salt and drought tolerance in rice.
|
| |
J Integr Plant Biol,
51,
825-833.
|
 |
|
|
|
|
 |
T.L.Arakaki,
F.S.Buckner,
J.R.Gillespie,
N.A.Malmquist,
M.A.Phillips,
O.Kalyuzhniy,
J.R.Luft,
G.T.Detitta,
C.L.Verlinde,
W.C.Van Voorhis,
W.G.Hol,
and
E.A.Merritt
(2008).
Characterization of Trypanosoma brucei dihydroorotate dehydrogenase as a possible drug target; structural, kinetic and RNAi studies.
|
| |
Mol Microbiol,
68,
37-50.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Zameitat,
A.J.Pierik,
K.Zocher,
and
M.Löffler
(2007).
Dihydroorotate dehydrogenase from Saccharomyces cerevisiae: spectroscopic investigations with the recombinant enzyme throw light on catalytic properties and metabolism of fumarate analogues.
|
| |
FEMS Yeast Res,
7,
897-904.
|
 |
|
|
|
|
 |
J.Shi,
J.Dertouzos,
A.Gafni,
D.Steel,
and
B.A.Palfey
(2006).
Single-molecule kinetics reveals signatures of half-sites reactivity in dihydroorotate dehydrogenase A catalysis.
|
| |
Proc Natl Acad Sci U S A,
103,
5775-5780.
|
 |
|
|
|
|
 |
M.Kilstrup,
K.Hammer,
P.Ruhdal Jensen,
and
J.Martinussen
(2005).
Nucleotide metabolism and its control in lactic acid bacteria.
|
| |
FEMS Microbiol Rev,
29,
555-590.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

