 |
PDBsum entry 1j4b

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.4
- adenylosuccinate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + L-aspartate + GTP = N6-(1,2-dicarboxyethyl)-AMP + GDP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
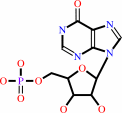 IMP
IMP
|
+
|
 L-aspartate
L-aspartate
|
+
|
 GTP
GTP
|
=
|
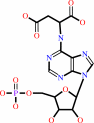 N(6)-(1,2-dicarboxyethyl)-AMP
N(6)-(1,2-dicarboxyethyl)-AMP
|
+
|
 GDP
GDP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
276:42146-42152
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Recombinant mouse muscle adenylosuccinate synthetase: overexpression, kinetics, and crystal structure.
|
|
C.V.Iancu,
T.Borza,
J.Y.Choe,
H.J.Fromm,
R.B.Honzatko.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Vertebrates possess two isozymes of adenylosuccinate synthetase. The acidic
isozyme is similar to the synthetase from bacteria and plants, being involved in
the de novo biosynthesis of AMP, whereas the basic isozyme participates in the
purine nucleotide cycle. Reported here is the first instance of overexpression
and crystal structure determination of a basic isozyme of adenylosuccinate
synthetase. The recombinant mouse muscle enzyme purified to homogeneity in
milligram quantities exhibits a specific activity comparable with that of the
rat muscle enzyme isolated from tissue and K(m) parameters for GTP, IMP, and
l-aspartate (12, 45, and 140 microm, respectively) similar to those of the
enzyme from Escherichia coli. The mouse muscle and E. coli enzymes have similar
polypeptide folds, differing primarily in the conformation of loops, involved in
substrate recognition and stabilization of the transition state. Residues 65-68
of the muscle isozyme adopt a conformation not observed in any previous
synthetase structure. In its new conformation, segment 65-68 forms
intramolecular hydrogen bonds with residues essential for the recognition of IMP
and, in fact, sterically excludes IMP from the active site. Observed differences
in ligand recognition among adenylosuccinate synthetases may be due in part to
conformational variations in the IMP pocket of the ligand-free enzymes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Subunit interface of mouse muscle
adenylosuccinate synthetase. Top panel, overview of the subunits
in the dimer as viewed down its molecular 2-fold axis. Bottom
panel, a more detailed presentation of side chain interactions.
The viewing orientation is the same for the top and bottom
panels. This figure was drawn by MOLSCRIPT (47).
|
 |
Figure 4.
Fig. 4. Stereoview of segment 65-68 of the mouse muscle
synthetase. Top panel, residues 65-68 and associated omit
electron density contoured at 1  with a
cut-off radius of 1 Å. Middle and bottom panels,
conformational differences between residues 65-68 of the mouse
muscle synthetase (bold lines) and corresponding residues of the
ligand-free (middle panel) and ligated (bottom panel) E. coli
enzyme. The dashed lines represent donor-acceptor interactions. with a
cut-off radius of 1 Å. Middle and bottom panels,
conformational differences between residues 65-68 of the mouse
muscle synthetase (bold lines) and corresponding residues of the
ligand-free (middle panel) and ligated (bottom panel) E. coli
enzyme. The dashed lines represent donor-acceptor interactions.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2001,
276,
42146-42152)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.R.Jefferson,
T.P.Walsh,
and
G.J.Barton
(2008).
A comparison of SCOP and CATH with respect to domain-domain interactions.
|
| |
Proteins,
70,
54-62.
|
 |
|
|
|
|
 |
H.Sun,
N.Li,
X.Wang,
T.Chen,
L.Shi,
L.Zhang,
J.Wang,
T.Wan,
and
X.Cao
(2005).
Molecular cloning and characterization of a novel muscle adenylosuccinate synthetase, AdSSL1, from human bone marrow stromal cells.
|
| |
Mol Cell Biochem,
269,
85-94.
|
 |
|
|
|
|
 |
H.Y.Wen,
Y.Xia,
M.E.Young,
H.Taegtmeyer,
and
R.E.Kellems
(2002).
The adenylosuccinate synthetase-1 gene is activated in the hypertrophied heart.
|
| |
J Cell Mol Med,
6,
235-243.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

