 |
PDBsum entry 1itq

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.4.13.19
- membrane dipeptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an L-aminoacyl-L-amino acid + H2O = 2 an L-alpha-amino acid
|
 |
 |
 |
 |
 |
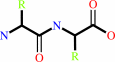 L-aminoacyl-L-amino acid
L-aminoacyl-L-amino acid
|
+
|
H2O
|
=
|
2
×
an L-alpha-amino acid
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.5.2.6
- beta-lactamase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a beta-lactam + H2O = a substituted beta-amino acid
|
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
321:177-184
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human renal dipeptidase involved in beta-lactam hydrolysis.
|
|
Y.Nitanai,
Y.Satow,
H.Adachi,
M.Tsujimoto.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human renal dipeptidase is a membrane-bound glycoprotein hydrolyzing dipeptides
and is involved in hydrolytic metabolism of penem and carbapenem beta-lactam
antibiotics. The crystal structures of the saccharide-trimmed enzyme are
determined as unliganded and inhibitor-liganded forms. They are informative for
designing new antibiotics that are not hydrolyzed by this enzyme. The active
site in each of the (alpha/beta)(8) barrel subunits of the homodimeric molecule
is composed of binuclear zinc ions bridged by the Glu125 side-chain located at
the bottom of the barrel, and it faces toward the microvillar membrane of a
kidney tubule. A dipeptidyl moiety of the therapeutically used cilastatin
inhibitor is fully accommodated in the active-site pocket, which is small enough
for precise recognition of dipeptide substrates. The barrel and active-site
architectures utilizing catalytic metal ions exhibit unexpected similarities to
those of the murine adenosine deaminase and the catalytic domain of the
bacterial urease.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The overall view of the hrDP. (a) The dimeric
structure of the hrDP viewed from the membrane-binding side.
Ser369 anchored to the membrane is located on the C-terminal
end, and the active sites are located on this side. The
a-helices and the b-strands composing (a/b)[8] barrels are shown
in green and yellow, respectively, and the a-helices capping the
barrels are shown in magenta. Zinc ions are drawn as red
spheres; cysteine residues forming disulfide bonds are drawn as
yellow ball-and-sticks; N-linked N-acetylglucosamine molecules
are drawn as pink ball-and-sticks. (b) The dimer viewed from a
90° rotation along the long axis of the dimer. (c) Stereo
representation of a monomer subunit. (d) A drawing of the
folding of the monomer subunit. The strands or helices are drawn
to reflect their relative lengths. The color scheme and labels
are consistent with those in (a) to (c). All Figures except
Figure 2(a) and (c) were produced with MOLSCRIPT [30.] and
Raster3D. [31.]
|
 |
Figure 3.
Figure 3. Comparison of the architectures of the
metallo-hydrolases having disordered (a/b)[8] barrels. (a)
Arrangement of b-strands composing the barrels. Corresponding
main-chain atoms of the b-strands are superimposed by
least-squares fitting. The strands of subunit A of the hrDP are
shown as yellow arrows; urease in sky-blue, phosphotriesterase
in orange, and adenosine deaminase in red. The strands labeled
b1 through b8 are sequentially arranged from the N-terminal side
in each polypeptide. (b) Superimposition of the metal ions and
ligands through least-squares fitting. The metal ions and ligand
groups of the hrDP subunit A are drawn in yellow, urease in
sky-blue, and phosphotriesterase in orange. Carbamoyllysine
ligands are represented with Carb-K labels. The fittings in (a)
and (b) were calculated independently.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2002,
321,
177-184)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.A.Cummings,
T.T.Nguyen,
A.A.Fedorov,
P.Kolb,
C.Xu,
E.V.Fedorov,
B.K.Shoichet,
D.P.Barondeau,
S.C.Almo,
and
F.M.Raushel
(2010).
Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase .
|
| |
Biochemistry,
49,
611-622.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.O'Dwyer,
R.Razzaque,
X.Hu,
S.K.Hollingshead,
and
J.G.Wall
(2009).
Engineering of cysteine residues leads to improved production of a human dipeptidase enzyme in E. coli.
|
| |
Appl Biochem Biotechnol,
159,
178-190.
|
 |
|
|
|
|
 |
E.Ristoff,
and
A.Larsson
(2007).
Inborn errors in the metabolism of glutathione.
|
| |
Orphanet J Rare Dis,
2,
16.
|
 |
|
|
|
|
 |
R.S.Hall,
D.F.Xiang,
C.Xu,
and
F.M.Raushel
(2007).
N-Acetyl-D-glucosamine-6-phosphate deacetylase: substrate activation via a single divalent metal ion.
|
| |
Biochemistry,
46,
7942-7952.
|
 |
|
|
|
|
 |
M.Goto,
H.Hayashi,
I.Miyahara,
K.Hirotsu,
M.Yoshida,
and
T.Oikawa
(2006).
Crystal structures of nonoxidative zinc-dependent 2,6-dihydroxybenzoate (gamma-resorcylate) decarboxylase from Rhizobium sp. strain MTP-10005.
|
| |
J Biol Chem,
281,
34365-34373.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.H.Baek,
J.J.Song,
S.J.Kwon,
C.Park,
C.M.Jung,
and
M.H.Sung
(2004).
Characteristics of a new enantioselective thermostable dipeptidase from Brevibacillus borstelensis BCS-1 and its application to synthesis of a D-amino-acid-containing dipeptide.
|
| |
Appl Environ Microbiol,
70,
1570-1575.
|
 |
|
|
|
|
 |
F.Vincent,
D.Yates,
E.Garman,
G.J.Davies,
and
J.A.Brannigan
(2004).
The three-dimensional structure of the N-acetylglucosamine-6-phosphate deacetylase, NagA, from Bacillus subtilis: a member of the urease superfamily.
|
| |
J Biol Chem,
279,
2809-2816.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Bochtler,
S.G.Odintsov,
M.Marcyjaniak,
and
I.Sabala
(2004).
Similar active sites in lysostaphins and D-Ala-D-Ala metallopeptidases.
|
| |
Protein Sci,
13,
854-861.
|
 |
|
|
|
|
 |
W.L.Lai,
L.Y.Chou,
C.Y.Ting,
R.Kirby,
Y.C.Tsai,
A.H.Wang,
and
S.H.Liaw
(2004).
The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
|
| |
J Biol Chem,
279,
13962-13967.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

