 |
PDBsum entry 1i2d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.4
- sulfate adenylyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
sulfate + ATP + H+ = adenosine 5'-phosphosulfate + diphosphate
|
 |
 |
 |
 |
 |
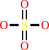 sulfate
sulfate
|
+
|
 ATP
ATP
|
+
|
H(+)
|
=
|
adenosine 5'-phosphosulfate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
40:6795-6804
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of ATP sulfurylase from Penicillium chrysogenum: insights into the allosteric regulation of sulfate assimilation.
|
|
I.J.MacRae,
I.H.Segel,
A.J.Fisher.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
ATP sulfurylase from Penicillium chrysogenum is an allosterically regulated
enzyme composed of six identical 63.7 kDa subunits (573 residues). The
C-terminal allosteric domain of each subunit is homologous to APS kinase. In the
presence of APS, the enzyme crystallized in the orthorhombic space group (I222)
with unit cell parameters of a = 135.7 A, b = 162.1 A, and c = 273.0 A. The
X-ray structure at 2.8 A resolution established that the hexameric enzyme is a
dimer of triads in the shape of an oblate ellipsoid 140 A diameter x 70 A. Each
subunit is divided into a discreet N-terminal domain, a central catalytic
domain, and a C-terminal allosteric domain. Two molecules of APS bound per
subunit clearly identify the catalytic and allosteric domains. The sequence
197QXRN200 is largely responsible for anchoring the phosphosulfate group of APS
at the active site of the catalytic domain. The specificity of the catalytic
site for adenine nucleotides is established by specific hydrogen bonds to the
protein main chain. APS was bound to the allosteric site through
sequence-specific interactions with amino acid side chains that are conserved in
true APS kinase. Within a given triad, the allosteric domain of one subunit
interacts with the catalytic domain of another. There are also
allosteric-allosteric, allosteric-N-terminal, and catalytic-catalytic domain
interactions across the triad interface. The overall interactions-each subunit
with four others-provide stability to the hexamer as well as a way to propagate
a concerted allosteric transition. The structure presented here is believed to
be the R state. A solvent channel, 15-70 A wide exists along the 3-fold axis,
but substrates have access to the catalytic site only from the external medium.
On the other hand, a surface "trench" links each catalytic site in one triad
with an allosteric site in the other triad. This trench may be a vestigial
feature of a bifunctional ("PAPS synthetase") ancestor of fungal ATP sulfurylase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.C.Gay,
I.H.Segel,
and
A.J.Fisher
(2009).
Structure of the two-domain hexameric APS kinase from Thiobacillus denitrificans: structural basis for the absence of ATP sulfurylase activity.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1021-1031.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.J.Patron,
D.G.Durnford,
and
S.Kopriva
(2008).
Sulfate assimilation in eukaryotes: fusions, relocations and lateral transfers.
|
| |
BMC Evol Biol,
8,
39.
|
 |
|
|
|
|
 |
O.Y.Gavel,
A.V.Kladova,
S.A.Bursakov,
J.M.Dias,
S.Texeira,
V.L.Shnyrov,
J.J.Moura,
I.Moura,
M.J.Romão,
and
J.Trincão
(2008).
Purification, crystallization and preliminary X-ray diffraction analysis of adenosine triphosphate sulfurylase (ATPS) from the sulfate-reducing bacterium Desulfovibrio desulfuricans ATCC 27774.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
593-595.
|
 |
|
|
|
|
 |
J.D.Mougous,
D.H.Lee,
S.C.Hubbard,
M.W.Schelle,
D.J.Vocadlo,
J.M.Berger,
and
C.R.Bertozzi
(2006).
Molecular basis for G protein control of the prokaryotic ATP sulfurylase.
|
| |
Mol Cell,
21,
109-122.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.W.Schelle,
and
C.R.Bertozzi
(2006).
Sulfate metabolism in mycobacteria.
|
| |
Chembiochem,
7,
1516-1524.
|
 |
|
|
|
|
 |
N.G.Richards,
and
M.S.Kilberg
(2006).
Asparagine synthetase chemotherapy.
|
| |
Annu Rev Biochem,
75,
629-654.
|
 |
|
|
|
|
 |
A.Hassibi,
C.Contag,
M.O.Vlad,
M.Hafezi,
T.H.Lee,
R.W.Davis,
and
N.Pourmand
(2005).
Bioluminescence regenerative cycle (BRC) system: theoretical considerations for nucleic acid quantification assays.
|
| |
Biophys Chem,
116,
175-185.
|
 |
|
|
|
|
 |
D.Mendoza-Cózatl,
H.Loza-Tavera,
A.Hernández-Navarro,
and
R.Moreno-Sánchez
(2005).
Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants.
|
| |
FEMS Microbiol Rev,
29,
653-671.
|
 |
|
|
|
|
 |
E.Hanna,
K.F.Ng,
I.J.MacRae,
C.J.Bley,
A.J.Fisher,
and
I.H.Segel
(2004).
Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory domain.
|
| |
J Biol Chem,
279,
4415-4424.
|
 |
|
|
|
|
 |
M.Gerstein,
and
N.Echols
(2004).
Exploring the range of protein flexibility, from a structural proteomics perspective.
|
| |
Curr Opin Chem Biol,
8,
14-19.
|
 |
|
|
|
|
 |
N.Fernandez-Fuentes,
A.Hermoso,
J.Espadaler,
E.Querol,
F.X.Aviles,
and
B.Oliva
(2004).
Classification of common functional loops of kinase super-families.
|
| |
Proteins,
56,
539-555.
|
 |
|
|
|
|
 |
S.Harjes,
A.Scheidig,
and
P.Bayer
(2004).
Expression, purification and crystallization of human 3'-phosphoadenosine-5'-phosphosulfate synthetase 1.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
350-352.
|
 |
|
|
|
|
 |
B.Singh,
and
N.B.Schwartz
(2003).
Identification and functional characterization of the novel BM-motif in the murine phosphoadenosine phosphosulfate (PAPS) synthetase.
|
| |
J Biol Chem,
278,
71-75.
|
 |
|
|
|
|
 |
Y.Taguchi,
J.Hoseki,
Y.Kakuta,
and
K.Fukuyama
(2003).
Overproduction, crystallization and preliminary X-ray diffraction analysis of probable ATP sulfurylase from Thermus thermophilus HB8.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1645-1647.
|
 |
|
|
|
|
 |
I.J.MacRae,
I.H.Segel,
and
A.J.Fisher
(2002).
Allosteric inhibition via R-state destabilization in ATP sulfurylase from Penicillium chrysogenum.
|
| |
Nat Struct Biol,
9,
945-949.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

