 |
PDBsum entry 1hdu

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Carboxypeptidase
|
PDB id
|
|

|

|
1hdu
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Carboxypeptidase
|
 |
|
Title:
|
 |
Crystal structure of bovine pancreatic carboxypeptidase a complexed with aminocarbonylphenylalanine at 1.75 a
|
|
Structure:
|
 |
Carboxypeptidase a. Chain: a, b, d, e. Ec: 3.4.17.1
|
|
Source:
|
 |
Bos bovis. Bovine. Organism_taxid: 9913. Organ: pancreas
|
|
Resolution:
|
 |
|
1.75Å
|
R-factor:
|
0.198
|
R-free:
|
0.229
|
|
|
Authors:
|
 |
J.H.Cho,N.-C.Ha,S.J.Chung,D.H.Kim,K.Y.Choi,B.-H.Oh
|
|
Key ref:
|
 |
J.H.Cho
et al.
(2002).
Insight into the stereochemistry in the inhibition of carboxypeptidase A with N-(hydroxyaminocarbonyl)phenylalanine: binding modes of an enantiomeric pair of the inhibitor to carboxypeptidase A.
Bioorg Med Chem Lett,
10,
2015-2022.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
17-Nov-00
|
Release date:
|
15-Nov-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00730
(CBPA1_BOVIN) -
Carboxypeptidase A1 from Bos taurus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
419 a.a.
307 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 9 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.4.17.1
- carboxypeptidase A.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Peptidyl-L-amino acid + H2O = peptide + L-amino acid
|
 |
 |
 |
 |
 |
|
+
|
|
=
|
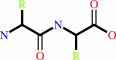
|
+
|
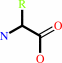
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Bioorg Med Chem Lett
10:2015-2022
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Insight into the stereochemistry in the inhibition of carboxypeptidase A with N-(hydroxyaminocarbonyl)phenylalanine: binding modes of an enantiomeric pair of the inhibitor to carboxypeptidase A.
|
|
J.H.Cho,
D.H.Kim,
S.J.Chung,
N.C.Ha,
B.H.Oh,
K.Yong Choi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Both D- and L-isomers of N-(hydroxyaminocarbonyl)phenylalanine () were shown to
have strong binding affinity towards carboxypeptidase A (CPA) with D- being more
potent than its enantiomer by 3-fold (Chung, S. J.; Kim, D. H. Bioorg. Med.
Chem. 2001, 9, 185.). In order to understand the reversed stereochemical
preference shown in the CPA inhibition, we have solved the crystal structures of
CPA complexed with each enantiometer of up to 1.75 A resolution. Inhibitor L-
whose stereochemistry belongs to the stereochemical series of substrate binds
CPA like substrate does with its carbonyl oxygen coordinating to the active site
zinc ion. Its hydroxyl is engaged in hydrogen bonding with the carboxylate of
Glu-270. On the other hand, in binding of D- to CPA, its terminal hydroxyl group
is involved in interactions with the active site zinc ion and the carboxylate of
Glu-270. In both CPA small middle dot complexes, the phenyl ring in is fitted in
the substrate recognition pocket at the S(1)' subsite, and the carboxylate of
the inhibitors forms bifurcated hydrogen bonds with the guanidinium moiety of
Arg-145 and a hydrogen bond with the guanidinium of Arg-127. In the complex of
CPA small middle dotD-, the carboxylate of the inhibitor is engaged in hydrogen
bonding with the phenolic hydroxyl of the down-positioned Tyr-248. While the L-
binding induces a concerted movement of the backbone amino acid residues at the
active site, only the downward movement of Tyr-248 was noted when D- binds to
CPA.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Haller,
P.Ekici,
A.Friess,
and
H.Parlar
(2010).
High enrichment of MMP-9 and carboxypeptidase A by tweezing adsorptive bubble separation (TABS).
|
| |
Appl Biochem Biotechnol,
162,
1547-1557.
|
 |
|
|
|
|
 |
D.Xu,
and
H.Guo
(2009).
Quantum mechanical/molecular mechanical and density functional theory studies of a prototypical zinc peptidase (carboxypeptidase A) suggest a general acid-general base mechanism.
|
| |
J Am Chem Soc,
131,
9780-9788.
|
 |
|
|
|
|
 |
K.H.Kim
(2007).
Outliers in SAR and QSAR: is unusual binding mode a possible source of outliers?
|
| |
J Comput Aided Mol Des,
21,
63-86.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

