 |
PDBsum entry 1h0c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.44
- alanine--glyoxylate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
glyoxylate + L-alanine = glycine + pyruvate
|
 |
 |
 |
 |
 |
glyoxylate
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
+
|
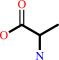 L-alanine
L-alanine
|
=
|
 glycine
glycine
|
+
|
 pyruvate
pyruvate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.51
- serine--pyruvate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-serine + pyruvate = 3-hydroxypyruvate + L-alanine
|
 |
 |
 |
 |
 |
 L-serine
L-serine
|
+
|
pyruvate
Bound ligand (Het Group name = )
matches with 71.43% similarity
|
=
|
3-hydroxypyruvate
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
+
|
 L-alanine
L-alanine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
331:643-652
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of alanine:glyoxylate aminotransferase and the relationship between genotype and enzymatic phenotype in primary hyperoxaluria type 1.
|
|
X.Zhang,
S.M.Roe,
Y.Hou,
M.Bartlam,
Z.Rao,
L.H.Pearl,
C.J.Danpure.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A deficiency of the liver-specific enzyme alanine:glyoxylate aminotransferase
(AGT) is responsible for the potentially lethal hereditary kidney stone disease
primary hyperoxaluria type 1 (PH1). Many of the mutations in the gene encoding
AGT are associated with specific enzymatic phenotypes such as accelerated
proteolysis (Ser205Pro), intra-peroxisomal aggregation (Gly41Arg), inhibition of
pyridoxal phosphate binding and loss of catalytic activity (Gly82Glu), and
peroxisome-to-mitochondrion mistargeting (Gly170Arg). Several mutations,
including that responsible for AGT mistargeting, co-segregate and interact
synergistically with a Pro11Leu polymorphism found at high frequency in the
normal population. In order to gain further insights into the mechanistic link
between genotype and enzymatic phenotype in PH1, we have determined the crystal
structure of normal human AGT complexed to the competitive inhibitor
amino-oxyacetic acid to 2.5A. Analysis of this structure allows the effects of
these mutations and polymorphism to be rationalised in terms of AGT tertiary and
quaternary conformation, and in particular it provides a possible explanation
for the Pro11Leu-Gly170Arg synergism that leads to AGT mistargeting.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Overall topology of the normal AGT dimer. (A) and
(B) The a-chain is colour-coded as follows: magenta, extended
N-terminal segment; red, large N-terminal domain; green, small
C-terminal domain. The b-chain is colour coded as follows: cyan,
extended N-terminal segment; yellow, large N-terminal domain;
blue, small C-terminal domain. B is rotated 180° in the
horizontal plane relative to A. Residues 41, 82, 170, 205, 209,
and 244 are indicated on the a-chain, residue 11 is indicated on
the b-chain. (C) Stereo pair of C^a chain, same orientation as
A. The pyridoxal phosphate cofactor is in shown in red and the
amino-oxyacetic acid inhibitor is shown in orange. The N and
C-terminal residues identified in the structure (4 and 390,
respectively) and every other 50 residues are numbered on the
a-chain (green).
|
 |
Figure 3.
Figure 3. Active site and dimerisation interface of normal
AGT. (A) Active site of AGT. AOA, amino-oxyacetic acid; Lys209a
is coloured magenta, the a-chain green, and the b-chain blue.
For PLP and AOA, the carbon atoms are in yellow, oxygen atoms in
red, phosphorous atom in orange, and nitrogen atoms in blue. Key
residues are labelled and important hydrogen bonds discussed in
the text are represented by dotted lines. (B) Locations of Gly41
and Gly42 at the dimerisation interface. The a-chain is coloured
green, and the b-chain blue.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2003,
331,
643-652)
copyright 2003.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Albert,
C.Yunta,
R.Arranz,
A.Peña,
E.Salido,
J.M.Valpuesta,
and
J.Martín-Benito
(2010).
Structure of GroEL in complex with an early folding intermediate of alanine glyoxylate aminotransferase.
|
| |
J Biol Chem,
285,
6371-6376.
|
 |
|
|
|
|
 |
B.Cellini,
R.Montioli,
A.Paiardini,
A.Lorenzetto,
F.Maset,
T.Bellini,
E.Oppici,
and
C.B.Voltattorni
(2010).
Molecular defects of the glycine 41 variants of alanine glyoxylate aminotransferase associated with primary hyperoxaluria type I.
|
| |
Proc Natl Acad Sci U S A,
107,
2896-2901.
|
 |
|
|
|
|
 |
I.Ramazzina,
R.Costa,
L.Cendron,
R.Berni,
A.Peracchi,
G.Zanotti,
and
R.Percudani
(2010).
An aminotransferase branch point connects purine catabolism to amino acid recycling.
|
| |
Nat Chem Biol,
6,
801-806.
|
 |
|
|
|
|
 |
R.N.Rodionov,
D.J.Murry,
S.F.Vaulman,
J.W.Stevens,
and
S.R.Lentz
(2010).
Human alanine-glyoxylate aminotransferase 2 lowers asymmetric dimethylarginine and protects from inhibition of nitric oxide production.
|
| |
J Biol Chem,
285,
5385-5391.
|
 |
|
|
|
|
 |
B.Cellini,
R.Montioli,
A.Paiardini,
A.Lorenzetto,
and
C.B.Voltattorni
(2009).
Molecular Insight into the Synergism between the Minor Allele of Human Liver Peroxisomal Alanine:Glyoxylate Aminotransferase and the F152I Mutation.
|
| |
J Biol Chem,
284,
8349-8358.
|
 |
|
|
|
|
 |
B.Hoppe,
B.B.Beck,
and
D.S.Milliner
(2009).
The primary hyperoxalurias.
|
| |
Kidney Int,
75,
1264-1271.
|
 |
|
|
|
|
 |
E.L.Williams,
C.Acquaviva,
A.Amoroso,
F.Chevalier,
M.Coulter-Mackie,
C.G.Monico,
D.Giachino,
T.Owen,
A.Robbiano,
E.Salido,
H.Waterham,
and
G.Rumsby
(2009).
Primary hyperoxaluria type 1: update and additional mutation analysis of the AGXT gene.
|
| |
Hum Mutat,
30,
910-917.
|
 |
|
|
|
|
 |
S.Donini,
M.Ferrari,
C.Fedeli,
M.Faini,
I.Lamberto,
A.S.Marletta,
L.Mellini,
M.Panini,
R.Percudani,
L.Pollegioni,
L.Caldinelli,
S.Petrucco,
and
A.Peracchi
(2009).
Recombinant production of eight human cytosolic aminotransferases and assessment of their potential involvement in glyoxylate metabolism.
|
| |
Biochem J,
422,
265-272.
|
 |
|
|
|
|
 |
E.D.Hopper,
A.M.Pittman,
M.C.Fitzgerald,
and
C.L.Tucker
(2008).
In Vivo and in Vitro Examination of Stability of Primary Hyperoxaluria-associated Human Alanine:Glyoxylate Aminotransferase.
|
| |
J Biol Chem,
283,
30493-30502.
|
 |
|
|
|
|
 |
F.Rossi,
R.Schwarcz,
and
M.Rizzi
(2008).
Curiosity to kill the KAT (kynurenine aminotransferase): structural insights into brain kynurenic acid synthesis.
|
| |
Curr Opin Struct Biol,
18,
748-755.
|
 |
|
|
|
|
 |
G.Rumsby
(2008).
Oxalate transport as contributor to primary hyperoxaluria: the jury is still out.
|
| |
Am J Kidney Dis,
52,
1031-1034.
|
 |
|
|
|
|
 |
H.Sakuraba,
K.Yoneda,
K.Takeuchi,
H.Tsuge,
N.Katunuma,
and
T.Ohshima
(2008).
Structure of an archaeal alanine:glyoxylate aminotransferase.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
696-699.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Ikeda,
H.Kanouchi,
and
Y.Minatogawa
(2008).
Characterization of peroxisomal targeting signals on alanine: glyoxylate aminotransferase.
|
| |
Biol Pharm Bull,
31,
131-134.
|
 |
|
|
|
|
 |
Y.Yoshikane,
N.Yokochi,
M.Yamasaki,
K.Mizutani,
K.Ohnishi,
B.Mikami,
H.Hayashi,
and
T.Yagi
(2008).
Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099.
|
| |
J Biol Chem,
283,
1120-1127.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Lima,
R.Khristoforov,
C.Momany,
and
R.S.Phillips
(2007).
Crystal structure of Homo sapiens kynureninase.
|
| |
Biochemistry,
46,
2735-2744.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.E.Bobrowski,
and
C.B.Langman
(2006).
Hyperoxaluria and systemic oxalosis: current therapy and future directions.
|
| |
Expert Opin Pharmacother,
7,
1887-1896.
|
 |
|
|
|
|
 |
D.Milliner
(2006).
Treatment of the primary hyperoxalurias: a new chapter.
|
| |
Kidney Int,
70,
1198-1200.
|
 |
|
|
|
|
 |
F.Rossi,
S.Garavaglia,
G.B.Giovenzana,
B.Arcà,
J.Li,
and
M.Rizzi
(2006).
Crystal structure of the Anopheles gambiae 3-hydroxykynurenine transaminase.
|
| |
Proc Natl Acad Sci U S A,
103,
5711-5716.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Han,
H.Robinson,
Y.G.Gao,
N.Vogelaar,
S.R.Wilson,
M.Rizzi,
and
J.Li
(2006).
Crystal structures of Aedes aegypti alanine glyoxylate aminotransferase.
|
| |
J Biol Chem,
281,
37175-37182.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Leumann,
and
B.Hoppe
(2005).
Primary hyperoxaluria type 1: is genotyping clinically helpful?
|
| |
Pediatr Nephrol,
20,
555-557.
|
 |
|
|
|
|
 |
G.W.Han,
R.Schwarzenbacher,
R.Page,
L.Jaroszewski,
P.Abdubek,
E.Ambing,
T.Biorac,
J.M.Canaves,
H.J.Chiu,
X.Dai,
A.M.Deacon,
M.DiDonato,
M.A.Elsliger,
A.Godzik,
C.Grittini,
S.K.Grzechnik,
J.Hale,
E.Hampton,
J.Haugen,
M.Hornsby,
H.E.Klock,
E.Koesema,
A.Kreusch,
P.Kuhn,
S.A.Lesley,
I.Levin,
D.McMullan,
T.M.McPhillips,
M.D.Miller,
A.Morse,
K.Moy,
E.Nigoghossian,
J.Ouyang,
J.Paulsen,
K.Quijano,
R.Reyes,
E.Sims,
G.Spraggon,
R.C.Stevens,
H.van den Bedem,
J.Velasquez,
J.Vincent,
F.von Delft,
X.Wang,
B.West,
A.White,
G.Wolf,
Q.Xu,
O.Zagnitko,
K.O.Hodgson,
J.Wooley,
and
I.A.Wilson
(2005).
Crystal structure of an alanine-glyoxylate aminotransferase from Anabaena sp. at 1.70 A resolution reveals a noncovalently linked PLP cofactor.
|
| |
Proteins,
58,
971-975.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.A.Huber,
G.M.Birdsey,
M.J.Lumb,
D.T.Prowse,
T.J.Perkins,
D.R.Knight,
and
C.J.Danpure
(2005).
Peroxisomal import of human alanine:glyoxylate aminotransferase requires ancillary targeting information remote from its C terminus.
|
| |
J Biol Chem,
280,
27111-27120.
|
 |
|
|
|
|
 |
S.Koul,
T.Johnson,
S.Pramanik,
and
H.Koul
(2005).
Cellular transfection to deliver alanine-glyoxylate aminotransferase to hepatocytes: a rational gene therapy for primary hyperoxaluria-1 (PH-1).
|
| |
Am J Nephrol,
25,
176-182.
|
 |
|
|
|
|
 |
C.B.Langman
(2004).
The molecular basis of kidney stones.
|
| |
Curr Opin Pediatr,
16,
188-193.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

