 |
PDBsum entry 1g0h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of mj0109 gene product inositol monophosphatase- fructose 1,6 bisphosphatase
|
|
Structure:
|
 |
Inositol monophosphatase. Chain: a, b. Synonym: mj0109 gene product. Engineered: yes
|
|
Source:
|
 |
Methanocaldococcus jannaschii. Organism_taxid: 2190. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.226
|
R-free:
|
0.279
|
|
|
Authors:
|
 |
K.A.Johnson,L.Chen,H.Yang,M.F.Roberts,B.Stec
|
Key ref:

|
 |
K.A.Johnson
et al.
(2001).
Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
Biochemistry,
40,
618-630.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
06-Oct-00
|
Release date:
|
14-Mar-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q57573
(BSUHB_METJA) -
Fructose-1,6-bisphosphatase/inositol-1-monophosphatase from Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
252 a.a.
252 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.11
- fructose-bisphosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pentose Phosphate Pathway (later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 1,6-bisphosphate + H2O = beta-D-fructose 6-phosphate + phosphate
|
 |
 |
 |
 |
 |
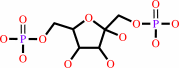 beta-D-fructose 1,6-bisphosphate
beta-D-fructose 1,6-bisphosphate
|
+
|
H2O
|
=
|
beta-D-fructose 6-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.3.25
- inositol-phosphate phosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a myo-inositol phosphate + H2O = myo-inositol + phosphate
|
 |
 |
 |
 |
 |
myo-inositol phosphate
|
+
|
H2O
|
=
|
myo-inositol
Bound ligand (Het Group name = )
matches with 75.00% similarity
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
40:618-630
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure and catalytic mechanism of the MJ0109 gene product: a bifunctional enzyme with inositol monophosphatase and fructose 1,6-bisphosphatase activities.
|
|
K.A.Johnson,
L.Chen,
H.Yang,
M.F.Roberts,
B.Stec.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Inositol monophosphatase (EC 3.1.3.25) in hyperthermophilic archaea is thought
to play a role in the biosynthesis of di-myo-inositol-1,1'-phosphate (DIP), an
osmolyte unique to hyperthermophiles. The Methanococcus jannaschii MJ109 gene
product, the sequence of which is substantially homologous to that of human
inositol monophosphatase, exhibits inositol monophosphatase activity but with
substrate specificity that is broader than those of bacterial and eukaryotic
inositol monophosphatases (it can also act as a fructose bisphosphatase). To
understand its substrate specificity as well as the poor inhibition by Li(+) (a
potent inhibitor of the mammalian enzyme), we have crystallized the enzyme and
determined its three-dimensional structure. The overall fold, as expected, is
similar to that of the mammalian enzyme, but the details suggest a closer
relationship to fructose 1,6-bisphosphatases. Three complexes of the MJ0109
protein with substrate and/or product and inhibitory as well as activating metal
ions suggest that the phosphatase mechanism is a three-metal ion assisted
catalysis which is in variance with that proposed previously for the human
inositol monophosphatase.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.Li,
K.A.Stieglitz,
A.L.Shrout,
Y.Wei,
R.M.Weis,
B.Stec,
and
M.F.Roberts
(2010).
Mobile loop mutations in an archaeal inositol monophosphatase: modulating three-metal ion assisted catalysis and lithium inhibition.
|
| |
Protein Sci,
19,
309-318.
|
 |
|
|
|
|
 |
G.Brown,
A.Singer,
V.V.Lunin,
M.Proudfoot,
T.Skarina,
R.Flick,
S.Kochinyan,
R.Sanishvili,
A.Joachimiak,
A.M.Edwards,
A.Savchenko,
and
A.F.Yakunin
(2009).
Structural and biochemical characterization of the type II fructose-1,6-bisphosphatase GlpX from Escherichia coli.
|
| |
J Biol Chem,
284,
3784-3792.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Kawai,
and
K.Murata
(2008).
Structure and function of NAD kinase and NADP phosphatase: key enzymes that regulate the intracellular balance of NAD(H) and NADP(H).
|
| |
Biosci Biotechnol Biochem,
72,
919-930.
|
 |
|
|
|
|
 |
A.K.Brown,
G.Meng,
H.Ghadbane,
D.J.Scott,
L.G.Dover,
J.Nigou,
G.S.Besra,
and
K.Fütterer
(2007).
Dimerization of inositol monophosphatase Mycobacterium tuberculosis SuhB is not constitutive, but induced by binding of the activator Mg2+.
|
| |
BMC Struct Biol,
7,
55.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Fukuda,
S.Kawai,
and
K.Murata
(2007).
NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3'-phosphoadenosine 5'-phosphate phosphatase.
|
| |
Appl Environ Microbiol,
73,
5447-5452.
|
 |
|
|
|
|
 |
K.A.Stieglitz,
M.F.Roberts,
W.Li,
and
B.Stec
(2007).
Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
|
| |
FEBS J,
274,
2461-2469.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Wang,
K.A.Stieglitz,
M.Bubunenko,
D.L.Court,
B.Stec,
and
M.F.Roberts
(2007).
The structure of the R184A mutant of the inositol monophosphatase encoded by suhB and implications for its functional interactions in Escherichia coli.
|
| |
J Biol Chem,
282,
26989-26996.
|
 |
|
|
|
|
 |
M.Goenrich,
R.K.Thauer,
H.Yurimoto,
and
N.Kato
(2005).
Formaldehyde activating enzyme (Fae) and hexulose-6-phosphate synthase (Hps) in Methanosarcina barkeri: a possible function in ribose-5-phosphate biosynthesis.
|
| |
Arch Microbiol,
184,
41-48.
|
 |
|
|
|
|
 |
R.Gill,
F.Mohammed,
R.Badyal,
L.Coates,
P.Erskine,
D.Thompson,
J.Cooper,
M.Gore,
and
S.Wood
(2005).
High-resolution structure of myo-inositol monophosphatase, the putative target of lithium therapy.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
545-555.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Nishimasu,
S.Fushinobu,
H.Shoun,
and
T.Wakagi
(2004).
The first crystal structure of the novel class of fructose-1,6-bisphosphatase present in thermophilic archaea.
|
| |
Structure,
12,
949-959.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.W.Nelson,
R.B.Honzatko,
and
H.J.Fromm
(2004).
Origin of cooperativity in the activation of fructose-1,6-bisphosphatase by Mg2+.
|
| |
J Biol Chem,
279,
18481-18487.
|
 |
|
|
|
|
 |
D.G.Kehres,
and
M.E.Maguire
(2003).
Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria.
|
| |
FEMS Microbiol Rev,
27,
263-290.
|
 |
|
|
|
|
 |
K.A.Stieglitz,
B.A.Seaton,
J.F.Head,
B.Stec,
and
M.F.Roberts
(2003).
Unexpected similarity in regulation between an archaeal inositol monophosphatase/fructose bisphosphatase and chloroplast fructose bisphosphatase.
|
| |
Protein Sci,
12,
760-767.
|
 |
|
|
|
|
 |
R.S.Ronimus,
and
H.W.Morgan
(2003).
Distribution and phylogenies of enzymes of the Embden-Meyerhof-Parnas pathway from archaea and hyperthermophilic bacteria support a gluconeogenic origin of metabolism.
|
| |
Archaea,
1,
199-221.
|
 |
|
|
|
|
 |
C.H.Verhees,
J.Akerboom,
E.Schiltz,
W.M.de Vos,
and
J.van der Oost
(2002).
Molecular and biochemical characterization of a distinct type of fructose-1,6-bisphosphatase from Pyrococcus furiosus.
|
| |
J Bacteriol,
184,
3401-3405.
|
 |
|
|
|
|
 |
K.A.Stieglitz,
K.A.Johnson,
H.Yang,
M.F.Roberts,
B.A.Seaton,
J.F.Head,
and
B.Stec
(2002).
Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
|
| |
J Biol Chem,
277,
22863-22874.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

