 |
PDBsum entry 1euw

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.6.1.23
- dUTP diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dUTP + H2O = dUMP + diphosphate + H+
|
 |
 |
 |
 |
 |
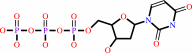 dUTP
dUTP
|
+
|
H2O
|
=
|
 dUMP
dUMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
57:767-774
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Atomic resolution structure of Escherichia coli dUTPase determined ab initio.
|
|
A.González,
G.Larsson,
R.Persson,
E.Cedergren-Zeppezauer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cryocooled crystals of a mercury complex of Escherichia coli dUTPase diffract to
atomic resolution. Data to 1.05 A resolution were collected from a derivative
crystal and the structure model was derived from a Fourier map with phases
calculated from the coordinates of the Hg atom (one site per subunit of the
trimeric enzyme) using the program ARP/wARP. After refinement with anisotropic
temperature factors a highly accurate model of the bacterial dUTPase was
obtained. Data to 1.45 A from a native crystal were also collected and the 100 K
structures were compared. Inspection of the refined models reveals that a large
part of the dUTPase remains rather mobile upon freezing, with 14% of the main
chain being totally disordered and with numerous side chains containing
disordered atoms in multiple discrete conformations. A large number of those
residues surround the active-site cavity. Two glycerol molecules (the
cryosolvent) occupy the deoxyribose-binding site. Comparison between the native
enzyme and the mercury complex shows that the active site is not adversely
affected by the binding of mercury. An unexpected effect seems to be a
stabilization of the crystal lattice by means of long-range interactions, making
derivatization a potentially useful tool for further studies of
inhibitor-substrate-analogue complexes of this protein at very high resolution.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4 Detail of the 1.05 Å structure, showing parts of the
polypeptide chain at the active site, including Tyr93 and one
molecule of glycerol (Glyc139). The 2mF[o] - DF[c]
electron-density map is contoured at 2  (in
blue) and 4 (in
blue) and 4  (coral). (coral).
|
 |
Figure 5.
Figure 5 View of the mercury-binding site in a 2mF[o] - DF[c]
electron-density map contoured at 3 and 10  (in
blue and orange, respectively) showing clearly the multiple
conformations of the Hg atom (red spheres) bound to the S (in
blue and orange, respectively) showing clearly the multiple
conformations of the Hg atom (red spheres) bound to the S  of
Cys36. of
Cys36.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2001,
57,
767-774)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.McNicholas,
E.Potterton,
K.S.Wilson,
and
M.E.Noble
(2011).
Presenting your structures: the CCP4mg molecular-graphics software.
|
| |
Acta Crystallogr D Biol Crystallogr,
67,
386-394.
|
 |
|
|
|
|
 |
J.García-Nafría,
L.Burchell,
M.Takezawa,
N.J.Rzechorzek,
M.J.Fogg,
and
K.S.Wilson
(2010).
The structure of the genomic Bacillus subtilis dUTPase: novel features in the Phe-lid.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
953-961.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.J.Davison,
and
N.D.Stow
(2005).
New genes from old: redeployment of dUTPase by herpesviruses.
|
| |
J Virol,
79,
12880-12892.
|
 |
|
|
|
|
 |
J.Kovári,
O.Barabás,
E.Takács,
A.Békési,
Z.Dubrovay,
V.Pongrácz,
I.Zagyva,
T.Imre,
P.Szabó,
and
B.G.Vértessy
(2004).
Altered active site flexibility and a structural metal-binding site in eukaryotic dUTPase: kinetic characterization, folding, and crystallographic studies of the homotrimeric Drosophila enzyme.
|
| |
J Biol Chem,
279,
17932-17944.
|
 |
|
|
|
|
 |
O.Barabás,
V.Pongrácz,
J.Kovári,
M.Wilmanns,
and
B.G.Vértessy
(2004).
Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUTPase.
|
| |
J Biol Chem,
279,
42907-42915.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Mustafi,
A.Bekesi,
B.G.Vertessy,
and
M.W.Makinen
(2003).
Catalytic and structural role of the metal ion in dUTP pyrophosphatase.
|
| |
Proc Natl Acad Sci U S A,
100,
5670-5675.
|
 |
|
|
|
|
 |
E.Johansson,
O.Bjornberg,
P.O.Nyman,
and
S.Larsen
(2003).
Structure of the bifunctional dCTP deaminase-dUTPase from Methanocaldococcus jannaschii and its relation to other homotrimeric dUTPases.
|
| |
J Biol Chem,
278,
27916-27922.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

