 |
PDBsum entry 1e2s

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.6.8
- cerebroside-sulfatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-acyl-1-beta-D-(3-O-sulfo)-galactosyl-sphing-4-enine + H2O = a beta- D-galactosyl-(1<->1')-N-acylsphing-4-enine + sulfate + H+
|
 |
 |
 |
 |
 |
N-acyl-1-beta-D-(3-O-sulfo)-galactosyl-sphing-4-enine
|
+
|
H2O
|
=
|
beta- D-galactosyl-(1<->1')-N-acylsphing-4-enine
|
+
|
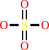 sulfate
sulfate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
305:269-277
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
|
|
R.von Bülow,
B.Schmidt,
T.Dierks,
K.von Figura,
I.Usón.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Arylsulfatase A (ASA) belongs to the sulfatase family whose members carry a
C(alpha)-formylglycine that is post-translationally generated by oxidation of a
conserved cysteine or serine residue. The crystal structures of two
arylsulfatases, ASA and ASB, and kinetic studies on ASA mutants led to different
proposals for the catalytic mechanism in the hydrolysis of sulfate esters. The
structures of two ASA mutants that lack the functional C(alpha)-formylglycine
residue 69, in complex with a synthetic substrate, have been determined in order
to unravel the reaction mechanism. The crystal structure of the inactive mutant
C69A-ASA in complex with p-nitrocatechol sulfate (pNCS) mimics a reaction
intermediate during sulfate ester hydrolysis by the active enzyme, without the
covalent bond to the key side-chain FGly69. The structure shows that the
side-chains of lysine 123, lysine 302, serine 150, histidine 229, the main-chain
of the key residue 69 and the divalent cation in the active center are involved
in sulfate binding. It is proposed that histidine 229 protonates the leaving
alcoholate after hydrolysis.C69S-ASA is able to bind covalently to the substrate
and hydrolyze it, but is unable to release the resulting sulfate. Nevertheless,
the resulting sulfation is low. The structure of C69S-ASA shows the serine
side-chain in a single conformation, turned away from the position a substrate
occupies in the complex. This suggests that the double conformation observed in
the structure of wild-type ASA is more likely to correspond to a formylglycine
hydrate than to a twofold disordered aldehyde oxo group, and accounts for the
relative inertness of the C69S-ASA mutant. In the C69S-ASA-pNCS complex, the
substrate occupies the same position as in the C69A-ASA-pNCS complex, which
corresponds to the non-covalently bonded substrate. Based on the structural
data, a detailed mechanism for sulfate ester cleavage is proposed, involving an
aldehyde hydrate as the functional group.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.R.Myette,
V.Soundararajan,
Z.Shriver,
R.Raman,
and
R.Sasisekharan
(2009).
Heparin/heparan sulfate 6-O-sulfatase from Flavobacterium heparinum: integrated structural and biochemical investigation of enzyme active site and substrate specificity.
|
| |
J Biol Chem,
284,
35177-35188.
|
 |
|
|
|
|
 |
M.Schenk,
C.A.Koppisetty,
D.C.Santos,
E.Carmona,
S.Bhatia,
P.G.Nyholm,
and
N.Tanphaichitr
(2009).
Interaction of arylsulfatase-A (ASA) with its natural sulfoglycolipid substrates: a computational and site-directed mutagenesis study.
|
| |
Glycoconj J,
26,
1029-1045.
|
 |
|
|
|
|
 |
T.S.Kang,
and
R.C.Stevens
(2009).
Structural aspects of therapeutic enzymes to treat metabolic disorders.
|
| |
Hum Mutat,
30,
1591-1610.
|
 |
|
|
|
|
 |
J.P.Lai,
J.R.Thompson,
D.S.Sandhu,
and
L.R.Roberts
(2008).
Heparin-degrading sulfatases in hepatocellular carcinoma: roles in pathogenesis and therapy targets.
|
| |
Future Oncol,
4,
803-814.
|
 |
|
|
|
|
 |
M.Mariappan,
K.Radhakrishnan,
T.Dierks,
B.Schmidt,
and
K.von Figura
(2008).
ERp44 mediates a thiol-independent retention of formylglycine-generating enzyme in the endoplasmic reticulum.
|
| |
J Biol Chem,
283,
6375-6383.
|
 |
|
|
|
|
 |
P.Bojarová,
E.Denehy,
I.Walker,
K.Loft,
D.P.De Souza,
L.W.Woo,
B.V.Potter,
M.J.McConville,
and
S.J.Williams
(2008).
Direct evidence for ArO-S bond cleavage upon inactivation of Pseudomonas aeruginosa arylsulfatase by aryl sulfamates.
|
| |
Chembiochem,
9,
613-623.
|
 |
|
|
|
|
 |
S.L.Gande,
M.Mariappan,
B.Schmidt,
T.H.Pringle,
K.von Figura,
and
T.Dierks
(2008).
Paralog of the formylglycine-generating enzyme--retention in the endoplasmic reticulum by canonical and noncanonical signals.
|
| |
FEBS J,
275,
1118-1130.
|
 |
|
|
|
|
 |
S.R.Hanson,
L.J.Whalen,
and
C.H.Wong
(2006).
Synthesis and evaluation of general mechanism-based inhibitors of sulfatases based on (difluoro)methyl phenyl sulfate and cyclic phenyl sulfamate motifs.
|
| |
Bioorg Med Chem,
14,
8386-8395.
|
 |
|
|
|
|
 |
A.Preusser-Kunze,
M.Mariappan,
B.Schmidt,
S.L.Gande,
K.Mutenda,
D.Wenzel,
K.von Figura,
and
T.Dierks
(2005).
Molecular characterization of the human Calpha-formylglycine-generating enzyme.
|
| |
J Biol Chem,
280,
14900-14910.
|
 |
|
|
|
|
 |
C.Lingwood,
M.Mylvaganam,
F.Minhas,
B.Binnington,
D.R.Branch,
and
R.Pomès
(2005).
The sulfogalactose moiety of sulfoglycosphingolipids serves as a mimic of tyrosine phosphate in many recognition processes. Prediction and demonstration of Src homology 2 domain/sulfogalactose binding.
|
| |
J Biol Chem,
280,
12542-12547.
|
 |
|
|
|
|
 |
C.T.Walsh,
S.Garneau-Tsodikova,
and
G.J.Gatto
(2005).
Protein posttranslational modifications: the chemistry of proteome diversifications.
|
| |
Angew Chem Int Ed Engl,
44,
7342-7372.
|
 |
|
|
|
|
 |
M.Mariappan,
A.Preusser-Kunze,
M.Balleininger,
N.Eiselt,
B.Schmidt,
S.L.Gande,
D.Wenzel,
T.Dierks,
and
K.von Figura
(2005).
Expression, localization, structural, and functional characterization of pFGE, the paralog of the Calpha-formylglycine-generating enzyme.
|
| |
J Biol Chem,
280,
15173-15179.
|
 |
|
|
|
|
 |
S.R.Wallner,
B.M.Nestl,
and
K.Faber
(2005).
Highly enantioselective stereo-inverting sec-alkylsulfatase activity of hyperthermophilic Archaea.
|
| |
Org Biomol Chem,
3,
2652-2656.
|
 |
|
|
|
|
 |
S.R.Wallner,
M.Bauer,
C.Würdemann,
P.Wecker,
F.O.Glöckner,
and
K.Faber
(2005).
Highly enantioselective sec-alkyl sulfatase activity of the marine planctomycete Rhodopirellula baltica shows retention of configuration.
|
| |
Angew Chem Int Ed Engl,
44,
6381-6384.
|
 |
|
|
|
|
 |
Q.Fang,
J.Peng,
and
T.Dierks
(2004).
Post-translational formylglycine modification of bacterial sulfatases by the radical S-adenosylmethionine protein AtsB.
|
| |
J Biol Chem,
279,
14570-14578.
|
 |
|
|
|
|
 |
S.R.Hanson,
M.D.Best,
and
C.H.Wong
(2004).
Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility.
|
| |
Angew Chem Int Ed Engl,
43,
5736-5763.
|
 |
|
|
|
|
 |
C.Marquordt,
Q.Fang,
E.Will,
J.Peng,
K.von Figura,
and
T.Dierks
(2003).
Posttranslational modification of serine to formylglycine in bacterial sulfatases. Recognition of the modification motif by the iron-sulfur protein AtsB.
|
| |
J Biol Chem,
278,
2212-2218.
|
 |
|
|
|
|
 |
J.Peng,
B.Schmidt,
K.von Figura,
and
T.Dierks
(2003).
Identification of formylglycine in sulfatases by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
|
| |
J Mass Spectrom,
38,
80-86.
|
 |
|
|
|
|
 |
R.Raman,
J.R.Myette,
Z.Shriver,
K.Pojasek,
G.Venkataraman,
and
R.Sasisekharan
(2003).
The heparin/heparan sulfate 2-O-sulfatase from Flavobacterium heparinum. A structural and biochemical study of the enzyme active site and saccharide substrate specificity.
|
| |
J Biol Chem,
278,
12167-12174.
|
 |
|
|
|
|
 |
T.Dierks,
B.Schmidt,
L.V.Borissenko,
J.Peng,
A.Preusser,
M.Mariappan,
and
K.von Figura
(2003).
Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(alpha)-formylglycine generating enzyme.
|
| |
Cell,
113,
435-444.
|
 |
|
|
|
|
 |
I.Boltes,
H.Czapinska,
A.Kahnert,
R.von Bülow,
T.Dierks,
B.Schmidt,
K.von Figura,
M.A.Kertesz,
and
I.Usón
(2001).
1.3 A structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family.
|
| |
Structure,
9,
483-491.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Fey,
M.Balleininger,
L.V.Borissenko,
B.Schmidt,
K.von Figura,
and
T.Dierks
(2001).
Characterization of posttranslational formylglycine formation by luminal components of the endoplasmic reticulum.
|
| |
J Biol Chem,
276,
47021-47028.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

