 |
PDBsum entry 1e1f

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of a monocot (maize zmglu1) beta-glucosidase in complex with p-nitrophenyl-beta-d-thioglucoside
|
|
Structure:
|
 |
Beta-glucosidase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Zea mays. Maize. Organism_taxid: 4577. Strain: cv. Mutin. Tissue: coleoptile. Organelle: chloroplast. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_cell: plys s cells.
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.199
|
R-free:
|
0.252
|
|
|
Authors:
|
 |
M.Czjzek,M.Cicek,D.R.Bevan,B.Henrissat,A.Esen
|
|
Key ref:
|
 |
M.Czjzek
et al.
(2001).
Crystal structure of a monocotyledon (maize ZMGlu1) beta-glucosidase and a model of its complex with p-nitrophenyl beta-D-thioglucoside.
Biochem J,
354,
37-46.
PubMed id:
|
 |
|
Date:
|
 |
|
03-May-00
|
Release date:
|
19-Feb-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P49235
(HGGL1_MAIZE) -
4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside beta-D-glucosidase 1, chloroplastic from Zea mays
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
566 a.a.
490 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.2.1.182
- 4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
DIMBOA beta-D-glucoside + H2O = DIMBOA + D-glucose
|
|
2.
|
DIBOA beta-D-glucoside + H2O = DIBOA + D-glucose
|
|
 |
 |
 |
 |
 |
DIMBOA beta-D-glucoside
|
+
|
H2O
|
=
|
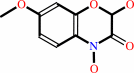 DIMBOA
DIMBOA
|
+
|
D-glucose
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
|
 |
 |
 |
 |
 |
DIBOA beta-D-glucoside
|
+
|
H2O
|
=
|
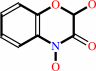 DIBOA
DIBOA
|
+
|
D-glucose
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.2.1.21
- beta-glucosidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Hydrolysis of terminal, non-reducing beta-D-glucose residues with release of beta-D-glucose.
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochem J
354:37-46
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of a monocotyledon (maize ZMGlu1) beta-glucosidase and a model of its complex with p-nitrophenyl beta-D-thioglucoside.
|
|
M.Czjzek,
M.Cicek,
V.Zamboni,
W.P.Burmeister,
D.R.Bevan,
B.Henrissat,
A.Esen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The maize beta-glucosidase isoenzymes ZMGlu1 and ZMGlu2 hydrolyse the abundant
natural substrate DIMBOAGlc
(2-O-beta-D-glucopyranosyl-4-hydroxy-7-methoxy-1,4-benzoxazin-3-one), whose
aglycone DIMBOA (2,4-hydroxy-7-methoxy-1,4-benzoxazin-3-one) is the major
defence chemical protecting seedlings and young plant parts against herbivores
and other pests. The two isoenzymes hydrolyse DIMBOAGlc with similar kinetics
but differ from each other and their sorghum homologues with respect to
specificity towards other substrates. To gain insights into the mechanism of
substrate (i.e. aglycone) specificity between the two maize isoenzymes and their
sorghum homologues, ZMGlu1 was produced in Escherichia coli, purified,
crystallized and its structure solved at 2.5 Angstrom resolution by X-ray
crystallography. In addition, the complex of ZMGlu1 with the non-hydrolysable
inhibitor p-nitrophenyl beta-D-thioglucoside was crystallized and, based on the
partial electron density, a model for the inhibitor molecule within the active
site is proposed. The inhibitor is located in a slot-like active site where its
aromatic aglycone is held by stacking interactions with Trp-378. Whereas some of
the atoms on the non-reducing end of the glucose moiety can be modelled on the
basis of the electron density, most of the inhibitor atoms are highly
disordered. This is attributed to the requirement of the enzyme to accommodate
two different species, namely the substrate in its ground state and in its
distorted conformation, for catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Nagao,
N.Nagano,
and
K.Mizuguchi
(2010).
Relationships between functional subclasses and information contained in active-site and ligand-binding residues in diverse superfamilies.
|
| |
Proteins,
78,
2369-2384.
|
 |
|
|
|
|
 |
J.R.Ketudat Cairns,
and
A.Esen
(2010).
β-Glucosidases.
|
| |
Cell Mol Life Sci,
67,
3389-3405.
|
 |
|
|
|
|
 |
A.Kumar,
N.K.Singhal,
B.Ramanujam,
A.Mitra,
N.R.Rameshwaram,
S.K.Nadimpalli,
and
C.P.Rao
(2009).
C(1)-/C(2)-aromatic-imino-glyco-conjugates: experimental and computational studies of binding, inhibition and docking aspects towards glycosidases isolated from soybean and jack bean.
|
| |
Glycoconj J,
26,
495-510.
|
 |
|
|
|
|
 |
A.D.Hill,
and
P.J.Reilly
(2008).
Computational analysis of glycoside hydrolase family 1 specificities.
|
| |
Biopolymers,
89,
1021-1031.
|
 |
|
|
|
|
 |
L.M.Mendonça,
and
S.R.Marana
(2008).
The role in the substrate specificity and catalysis of residues forming the substrate aglycone-binding site of a beta-glycosidase.
|
| |
FEBS J,
275,
2536-2547.
|
 |
|
|
|
|
 |
R.Dopitová,
P.Mazura,
L.Janda,
R.Chaloupková,
P.Jerábek,
J.Damborský,
T.Filipi,
N.S.Kiran,
and
B.Brzobohatý
(2008).
Functional analysis of the aglycone-binding site of the maize beta-glucosidase Zm-p60.1.
|
| |
FEBS J,
275,
6123-6135.
|
 |
|
|
|
|
 |
M.León,
P.Isorna,
M.Menéndez,
J.Sanz-Aparicio,
and
J.Polaina
(2007).
Comparative study and mutational analysis of distinctive structural elements of hyperthermophilic enzymes.
|
| |
Protein J,
26,
435-444.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Krause,
and
E.Kothe
(2006).
Use of RNA fingerprinting to identify fungal genes specifically expressed during ectomycorrhizal interaction.
|
| |
J Basic Microbiol,
46,
387-399.
|
 |
|
|
|
|
 |
X.Ma,
J.Koepke,
A.Bayer,
G.Fritzsch,
H.Michel,
and
J.Stöckigt
(2005).
Crystallization and preliminary X-ray analysis of native and selenomethionyl vinorine synthase from Rauvolfia serpentina.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
694-696.
|
 |
|
|
|
|
 |
J.K.McCarthy,
A.Uzelac,
D.F.Davis,
and
D.E.Eveleigh
(2004).
Improved catalytic efficiency and active site modification of 1,4-beta-D-glucan glucohydrolase A from Thermotoga neapolitana by directed evolution.
|
| |
J Biol Chem,
279,
11495-11502.
|
 |
|
|
|
|
 |
L.Verdoucq,
J.Morinière,
D.R.Bevan,
A.Esen,
A.Vasella,
B.Henrissat,
and
M.Czjze
(2004).
Structural determinants of substrate specificity in family 1 beta-glucosidases: novel insights from the crystal structure of sorghum dhurrinase-1, a plant beta-glucosidase with strict specificity, in complex with its natural substrate.
|
| |
J Biol Chem,
279,
31796-31803.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Bismuto,
F.Febbraio,
S.Limongelli,
R.Briante,
and
R.Nucci
(2003).
Dynamic fluorescence studies of beta-glycosidase mutants from Sulfolobus solfataricus: effects of single mutations on protein thermostability.
|
| |
Proteins,
51,
10-20.
|
 |
|
|
|
|
 |
L.Verdoucq,
M.Czjzek,
J.Moriniere,
D.R.Bevan,
and
A.Esen
(2003).
Mutational and structural analysis of aglycone specificity in maize and sorghum beta-glucosidases.
|
| |
J Biol Chem,
278,
25055-25062.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Wang,
X.He,
S.Yang,
X.An,
W.Chang,
and
D.Liang
(2003).
Structural basis for thermostability of beta-glycosidase from the thermophilic eubacterium Thermus nonproteolyticus HG102.
|
| |
J Bacteriol,
185,
4248-4255.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Kaper,
H.H.van Heusden,
B.van Loo,
A.Vasella,
J.van der Oost,
and
W.M.de Vos
(2002).
Substrate specificity engineering of beta-mannosidase and beta-glucosidase from Pyrococcus by exchange of unique active site residues.
|
| |
Biochemistry,
41,
4147-4155.
|
 |
|
|
|
|
 |
Y.Bhatia,
S.Mishra,
and
V.S.Bisaria
(2002).
Microbial beta-glucosidases: cloning, properties, and applications.
|
| |
Crit Rev Biotechnol,
22,
375-407.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

