 |
PDBsum entry 1d0s

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.21
- nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Corrin Biosynthesis (part 8)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5,6-dimethylbenzimidazole + nicotinate beta-D-ribonucleotide = alpha- ribazole 5'-phosphate + nicotinate + H+
|
 |
 |
 |
 |
 |
5,6-dimethylbenzimidazole
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
nicotinate beta-D-ribonucleotide
|
=
|
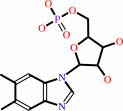 alpha- ribazole 5'-phosphate
alpha- ribazole 5'-phosphate
|
+
|
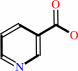 nicotinate
nicotinate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
38:16125-16135
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
The three-dimensional structures of nicotinate mononucleotide:5,6- dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella typhimurium complexed with 5,6-dimethybenzimidazole and its reaction products determined to 1.9 A resolution.
|
|
C.G.Cheong,
J.C.Escalante-Semerena,
I.Rayment.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase
(CobT) from Salmonella typhimurium plays a central role in the synthesis of
alpha-ribazole, which is a key component of the lower ligand of cobalamin. Two
X-ray structures of CobT are reported here at 1.9 A resolution. First, a complex
of CobT with 5,6-dimethylbenzimidazole, and second, a complex of CobT with its
reaction products, nicotinate and alpha-ribazole-5'-phosphate. CobT was
cocrystallized with 5,6-dimethylbenzimidazole (DMB) in the space group
P2(1)2(1)2 with unit cell dimensions of a = 72.1 A, b = 90.2 A, and c = 47.5 A
and one protomer per asymmetric unit. Subsequently, the crystals containing DMB
were soaked in nicotinate mononucleotide whereupon the physiological reaction
occurred in the crystal lattice to yield nicotinate and
alpha-ribazole-5'-phosphate. These studies show that CobT is a dimer where each
subunit consists of two domains. The large domain is dominated by a parallel
six-stranded beta-sheet with connecting alpha-helices that exhibit the topology
of a Rossmann fold. The small domain is made from components of the N- and
C-terminal sections of the polypeptide chain and contains a three-helix bundle.
The fold of CobT is unrelated to the type I and II phosphoribosylpyrophosphate
dependent transferases and does not appear to be related to any other protein
whose structure is known. The enzyme active site is located in a large cavity
formed by the loops at the C-terminal ends of the beta-strands and the small
domain of the neighboring subunit. DMB binds in a hydrophobic pocket created in
part by the neighboring small domain. This is consistent with the broad
specificity of this enzyme for aromatic substrates [Trzebiatowski, J. R.,
Escalante-Semerena (1997) J. Biol. Chem. 272, 17662-17667]. The binding site for
DMB suggests that Glu317 is the catalytic base required for the reaction. The
remainder of the cavity binds the nicotinate and ribose-5'-phosphate moieties,
which are nestled within the loops at the ends of the beta-strands.
Interestingly, the orientation of the substrate and products are opposite from
that expected for a Rossmann fold.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.R.Claas,
J.R.Parrish,
L.A.Maggio-Hall,
and
J.C.Escalante-Semerena
(2010).
Functional analysis of the nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) enzyme, involved in the late steps of coenzyme B12 biosynthesis in Salmonella enterica.
|
| |
J Bacteriol,
192,
145-154.
|
 |
|
|
|
|
 |
C.L.Zayas,
and
J.C.Escalante-Semerena
(2007).
Reassessment of the late steps of coenzyme B12 synthesis in Salmonella enterica: evidence that dephosphorylation of adenosylcobalamin-5'-phosphate by the CobC phosphatase is the last step of the pathway.
|
| |
J Bacteriol,
189,
2210-2218.
|
 |
|
|
|
|
 |
J.C.Escalante-Semerena
(2007).
Conversion of cobinamide into adenosylcobamide in bacteria and archaea.
|
| |
J Bacteriol,
189,
4555-4560.
|
 |
|
|
|
|
 |
C.G.Cheong,
J.C.Escalante-Semerena,
and
I.Rayment
(2002).
Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
|
| |
J Biol Chem,
277,
41120-41127.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

