 |
PDBsum entry 1cib

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.4
- adenylosuccinate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + L-aspartate + GTP = N6-(1,2-dicarboxyethyl)-AMP + GDP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
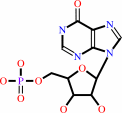 IMP
IMP
|
+
|
 L-aspartate
L-aspartate
|
+
|
GTP
Bound ligand (Het Group name = )
matches with 41.67% similarity
|
=
|
N(6)-(1,2-dicarboxyethyl)-AMP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 GDP
GDP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
274:17505-17510
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase.
|
|
Z.Hou,
M.Cashel,
H.J.Fromm,
R.B.Honzatko.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Guanosine 5'-diphosphate 3'-diphosphate (ppGpp), a pleiotropic effector of the
stringent response, potently inhibits adenylosuccinate synthetase from
Escherichia coli as an allosteric effector and/or as a competitive inhibitor
with respect to GTP. Crystals of the synthetase grown in the presence of IMP,
hadacidin, NO3-, and Mg2+, then soaked with ppGpp, reveal electron density at
the GTP pocket which is consistent with guanosine 5'-diphosphate 2':3'-cyclic
monophosphate. Unlike ligand complexes of the synthetase involving IMP and GDP,
the coordination of Mg2+ in this complex is octahedral with the side chain of
Asp13 in the inner sphere of the cation. The cyclic phosphoryl group interacts
directly with the side chain of Lys49 and indirectly through bridging water
molecules with the side chains of Asn295 and Arg305. The synthetase either
directly facilitates the formation of the cyclic nucleotide or scavenges trace
amounts of the cyclic nucleotide from solution. Regardless of its mode of
generation, the cyclic nucleotide binds far more tightly to the active site than
does ppGpp. Conceivably, synthetase activity in vivo during the stringent
response may be sensitive to the relative concentrations of several effectors,
which together exercise precise control over the de novo synthesis of AMP.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Schematic of ppG2':3'p showing atom labels.
|
 |
Figure 3.
Fig. 3. Stereoview of interactions between ppG2':3'p and
the active site. Ligands (only hadacidin (Had), nitrate,
ppG2':3'p, and Mg^2+ are shown) are drawn with bold lines.
Donor-acceptor interactions (corresponding distances listed in
Table II) are presented with dashed lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(1999,
274,
17505-17510)
copyright 1999.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Podzelinska,
S.M.He,
M.Wathier,
A.Yakunin,
M.Proudfoot,
B.Hove-Jensen,
D.L.Zechel,
and
Z.Jia
(2009).
Structure of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway for phosphonate degradation.
|
| |
J Biol Chem,
284,
17216-17226.
|
 |
|
|
|
|
 |
M.Montero,
G.Eydallin,
A.M.Viale,
G.Almagro,
F.J.Muñoz,
M.Rahimpour,
M.T.Sesma,
E.Baroja-Fernández,
and
J.Pozueta-Romero
(2009).
Escherichia coli glycogen metabolism is controlled by the PhoP-PhoQ regulatory system at submillimolar environmental Mg2+ concentrations, and is highly interconnected with a wide variety of cellular processes.
|
| |
Biochem J,
424,
129-141.
|
 |
|
|
|
|
 |
Y.Natori,
K.Tagami,
K.Murakami,
S.Yoshida,
O.Tanigawa,
Y.Moh,
K.Masuda,
T.Wada,
S.Suzuki,
H.Nanamiya,
Y.Tozawa,
and
F.Kawamura
(2009).
Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient in (p)ppGpp synthetase genes, relA, yjbM, and ywaC.
|
| |
J Bacteriol,
191,
4555-4561.
|
 |
|
|
|
|
 |
K.Mizusawa,
S.Masuda,
and
H.Ohta
(2008).
Expression profiling of four RelA/SpoT-like proteins, homologues of bacterial stringent factors, in Arabidopsis thaliana.
|
| |
Planta,
228,
553-562.
|
 |
|
|
|
|
 |
S.Masuda,
K.Mizusawa,
T.Narisawa,
Y.Tozawa,
H.Ohta,
and
K.Takamiya
(2008).
The bacterial stringent response, conserved in chloroplasts, controls plant fertilization.
|
| |
Plant Cell Physiol,
49,
135-141.
|
 |
|
|
|
|
 |
T.Hogg,
U.Mechold,
H.Malke,
M.Cashel,
and
R.Hilgenfeld
(2004).
Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected].
|
| |
Cell,
117,
57-68.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Chatterji,
and
A.K.Ojha
(2001).
Revisiting the stringent response, ppGpp and starvation signaling.
|
| |
Curr Opin Microbiol,
4,
160-165.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

