 |
PDBsum entry 1c4x

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (bphd) from rhodococcus sp. Strain rha1
|
|
Structure:
|
 |
Protein (2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase). Chain: a. Synonym: bphd. Engineered: yes
|
|
Source:
|
 |
Rhodococcus sp.. Organism_taxid: 101510. Strain: rha1. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Octamer (from PDB file)
Octamer (from PDB file)
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.178
|
R-free:
|
0.227
|
|
|
Authors:
|
 |
N.Nandhagopal,T.Senda,Y.Mitsui
|
|
Key ref:
|
 |
N.Nandhagopal
et al.
(1997).
Three-Dimensional structure of microbial 2-Hydroxyl-6-Oxo-6-Phenylhexa-2,4- Dienoic acid (hpda) hydrolase (bphd enzyme) from rhodococcus sp. Strain rha1, In the pcb degradation pathway.
Proc jpn acad ,Ser b,
73,
154.
|
 |
|
Date:
|
 |
|
30-Sep-99
|
Release date:
|
01-Oct-99
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q75WN8
(Q75WN8_RHOJR) -
2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase from Rhodococcus jostii (strain RHA1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
285 a.a.
281 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.7.1.8
- 2,6-dioxo-6-phenylhexa-3-enoate hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2,6-dioxo-6-phenylhexa-3-enoate + H2O = 2-oxopent-4-enoate + benzoate + H+
|
 |
 |
 |
 |
 |
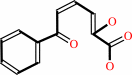 2,6-dioxo-6-phenylhexa-3-enoate
2,6-dioxo-6-phenylhexa-3-enoate
|
+
|
H2O
|
=
|
 2-oxopent-4-enoate
2-oxopent-4-enoate
|
+
|
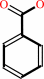 benzoate
benzoate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

