 |
PDBsum entry 1bvq

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.2.23
- 4-hydroxybenzoyl-CoA thioesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-hydroxybenzoyl-CoA + H2O = 4-hydroxybenzoate + CoA + H+
|
 |
 |
 |
 |
 |
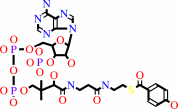 4-hydroxybenzoyl-CoA
4-hydroxybenzoyl-CoA
|
+
|
H2O
|
=
|
 4-hydroxybenzoate
4-hydroxybenzoate
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
273:33572-33579
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
The three-dimensional structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. Strain CBS-3.
|
|
M.M.Benning,
G.Wesenberg,
R.Liu,
K.L.Taylor,
D.Dunaway-Mariano,
H.M.Holden.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The soil-dwelling microbe, Pseudomonas sp. strain CBS-3, has attracted recent
attention due to its ability to survive on 4-chlorobenzoate as its sole carbon
source. The biochemical pathway by which this organism converts 4-chlorobenzoate
to 4-hydroxybenzoate consists of three enzymes: 4-chlorobenzoyl-CoA ligase,
4-chlorobenzoyl-CoA dehalogenase, and 4-hydroxybenzoyl-CoA thioesterase. Here we
describe the three-dimensional structure of the thioesterase determined to 2.0-A
resolution. Each subunit of the homotetramer is characterized by a five-stranded
anti-parallel beta-sheet and three major alpha-helices. While previous amino
acid sequence analyses failed to reveal any similarity between this thioesterase
and other known proteins, the results from this study clearly demonstrate that
the molecular architecture of 4-hydroxybenzoyl-CoA thioesterase is topologically
equivalent to that observed for beta-hydroxydecanoyl thiol ester dehydrase from
Escherichia coli. On the basis of the structural similarity between these two
enzymes, the active site of the thioesterase has been identified and a catalytic
mechanism proposed.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. Ribbon representation of one subunit of the
4-hydroxybenzoyl-CoA thioesterase. This figure and Figs. 3-5
were prepared with the software package, MOLSCRIPT (38). The
 -strands are
labeled A-F. -strands are
labeled A-F.
|
 |
Figure 4.
Fig. 4. Superposition of the  -carbons
for 4-hydroxybenzoyl-CoA thioesterase and -carbons
for 4-hydroxybenzoyl-CoA thioesterase and  -hydroxydecanoyl
thiol ester dehydrase. X-ray coordinates for the dehydrase were
obtained from the Brookhaven Protein Data Bank (1MKA). The
dehydrase and the thioesterase are displayed in red and black,
respectively. The 3-decynoyl-N-acetylcysteamine suicide
inhibitor observed in the dehydrase structure is depicted in a
ball-and-stick representation. -hydroxydecanoyl
thiol ester dehydrase. X-ray coordinates for the dehydrase were
obtained from the Brookhaven Protein Data Bank (1MKA). The
dehydrase and the thioesterase are displayed in red and black,
respectively. The 3-decynoyl-N-acetylcysteamine suicide
inhibitor observed in the dehydrase structure is depicted in a
ball-and-stick representation.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(1998,
273,
33572-33579)
copyright 1998.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.E.Alber
(2011).
Biotechnological potential of the ethylmalonyl-CoA pathway.
|
| |
Appl Microbiol Biotechnol,
89,
17-25.
|
 |
|
|
|
|
 |
D.C.Cantu,
Y.Chen,
and
P.J.Reilly
(2010).
Thioesterases: a new perspective based on their primary and tertiary structures.
|
| |
Protein Sci,
19,
1281-1295.
|
 |
|
|
|
|
 |
M.V.Dias,
F.Huang,
D.Y.Chirgadze,
M.Tosin,
D.Spiteller,
E.F.Dry,
P.F.Leadlay,
J.B.Spencer,
and
T.L.Blundell
(2010).
Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
|
| |
J Biol Chem,
285,
22495-22504.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.S.Pidugu,
K.Maity,
K.Ramaswamy,
N.Surolia,
and
K.Suguna
(2009).
Analysis of proteins with the 'hot dog' fold: prediction of function and identification of catalytic residues of hypothetical proteins.
|
| |
BMC Struct Biol,
9,
37.
|
 |
|
|
|
|
 |
M.Kotaka,
R.Kong,
I.Qureshi,
Q.S.Ho,
H.Sun,
C.W.Liew,
L.P.Goh,
P.Cheung,
Y.Mu,
J.Lescar,
and
Z.X.Liang
(2009).
Structure and catalytic mechanism of the thioesterase CalE7 in enediyne biosynthesis.
|
| |
J Biol Chem,
284,
15739-15749.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Awakawa,
K.Yokota,
N.Funa,
F.Doi,
N.Mori,
H.Watanabe,
and
S.Horinouchi
(2009).
Physically discrete beta-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase.
|
| |
Chem Biol,
16,
613-623.
|
 |
|
|
|
|
 |
T.Hosaka,
K.Murayama,
M.Kato-Murayama,
A.Urushibata,
R.Akasaka,
T.Terada,
M.Shirouzu,
S.Kuramitsu,
and
S.Yokoyama
(2009).
Structure of the putative thioesterase protein TTHA1846 from Thermus thermophilus HB8 complexed with coenzyme A and a zinc ion.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
767-776.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Yokoyama,
K.J.Choi,
A.M.Bosch,
and
H.J.Yeo
(2009).
Structure and function of a Campylobacter jejuni thioesterase Cj0915, a hexameric hot dog fold enzyme.
|
| |
Biochim Biophys Acta,
1794,
1073-1081.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Angelini,
L.Cendron,
S.Goncalves,
G.Zanotti,
and
L.Terradot
(2008).
Structural and enzymatic characterization of HP0496, a YbgC thioesterase from Helicobacter pylori.
|
| |
Proteins,
72,
1212-1221.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Geng,
J.B.Bruhn,
K.F.Nielsen,
L.Gram,
and
R.Belas
(2008).
Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters.
|
| |
Appl Environ Microbiol,
74,
1535-1545.
|
 |
|
|
|
|
 |
M.Chruszcz,
M.D.Zimmerman,
S.Wang,
K.D.Koclega,
H.Zheng,
E.Evdokimova,
M.Kudritska,
M.Cymborowski,
A.Savchenko,
A.Edwards,
and
W.Minor
(2008).
Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
|
| |
Cryst Growth Des,
8,
4054-4061.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Yokoyama,
S.Paek,
C.P.Ewing,
P.Guerry,
and
H.J.Yeo
(2008).
Structure of a sigma28-regulated nonflagellar virulence protein from Campylobacter jejuni.
|
| |
J Mol Biol,
384,
364-376.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.K.Forwood,
A.S.Thakur,
G.Guncar,
M.Marfori,
D.Mouradov,
W.Meng,
J.Robinson,
T.Huber,
S.Kellie,
J.L.Martin,
D.A.Hume,
and
B.Kobe
(2007).
Structural basis for recruitment of tandem hotdog domains in acyl-CoA thioesterase 7 and its role in inflammation.
|
| |
Proc Natl Acad Sci U S A,
104,
10382-10387.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Gully,
and
E.Bouveret
(2006).
A protein network for phospholipid synthesis uncovered by a variant of the tandem affinity purification method in Escherichia coli.
|
| |
Proteomics,
6,
282-293.
|
 |
|
|
|
|
 |
G.E.Schujman,
M.Guerin,
A.Buschiazzo,
F.Schaeffer,
L.I.Llarrull,
G.Reh,
A.J.Vila,
P.M.Alzari,
and
D.de Mendoza
(2006).
Structural basis of lipid biosynthesis regulation in Gram-positive bacteria.
|
| |
EMBO J,
25,
4074-4083.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.W.Giraldes,
D.L.Akey,
J.D.Kittendorf,
D.H.Sherman,
J.L.Smith,
and
R.A.Fecik
(2006).
Structural and mechanistic insights into polyketide macrolactonization from polyketide-based affinity labels.
|
| |
Nat Chem Biol,
2,
531-536.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.H.Chin,
C.C.Chou,
A.H.Wang,
and
S.H.Chou
(2006).
Crystal structure of a putative acyl-CoA thioesterase from Xanthomonas campestris (XC229) adopts a tetrameric hotdog fold of epsilongamma mode.
|
| |
Proteins,
64,
823-826.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Johansson,
A.Castell,
T.A.Jones,
and
K.Bäckbro
(2006).
Structure and function of Rv0130, a conserved hypothetical protein from Mycobacterium tuberculosis.
|
| |
Protein Sci,
15,
2300-2309.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Serek,
J.K.Forwood,
D.A.Hume,
J.L.Martin,
and
B.Kobe
(2006).
Crystallization of the C-terminal domain of the mouse brain cytosolic long-chain acyl-CoA thioesterase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
133-135.
|
 |
|
|
|
|
 |
D.B.Janssen,
I.J.Dinkla,
G.J.Poelarends,
and
P.Terpstra
(2005).
Bacterial degradation of xenobiotic compounds: evolution and distribution of novel enzyme activities.
|
| |
Environ Microbiol,
7,
1868-1882.
|
 |
|
|
|
|
 |
D.H.Pieper
(2005).
Aerobic degradation of polychlorinated biphenyls.
|
| |
Appl Microbiol Biotechnol,
67,
170-191.
|
 |
|
|
|
|
 |
D.Kostrewa,
F.K.Winkler,
G.Folkers,
L.Scapozza,
and
R.Perozzo
(2005).
The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum.
|
| |
Protein Sci,
14,
1570-1580.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Serrano-Vega,
R.Garcés,
and
E.Martínez-Force
(2005).
Cloning, characterization and structural model of a FatA-type thioesterase from sunflower seeds (Helianthus annuus L.).
|
| |
Planta,
221,
868-880.
|
 |
|
|
|
|
 |
A.Berchanski,
B.Shapira,
and
M.Eisenstein
(2004).
Hydrophobic complementarity in protein-protein docking.
|
| |
Proteins,
56,
130-142.
|
 |
|
|
|
|
 |
B.Sannigrahi,
P.McGeady,
and
I.M.Khan
(2004).
Helical poly(3-methyl-4-vinylpyridine)/amino acid complexes: preparation, characterization, and biocompatibility.
|
| |
Macromol Biosci,
4,
999.
|
 |
|
|
|
|
 |
S.C.Dillon,
and
A.Bateman
(2004).
The Hotdog fold: wrapping up a superfamily of thioesterases and dehydratases.
|
| |
BMC Bioinformatics,
5,
109.
|
 |
|
|
|
|
 |
Y.Tajika,
N.Sakai,
Y.Tanaka,
M.Yao,
N.Watanabe,
and
I.Tanaka
(2004).
Crystal structure of conserved protein PH1136 from Pyrococcus horikoshii.
|
| |
Proteins,
55,
210-213.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Berchanski,
and
M.Eisenstein
(2003).
Construction of molecular assemblies via docking: modeling of tetramers with D2 symmetry.
|
| |
Proteins,
53,
817-829.
|
 |
|
|
|
|
 |
M.Sosio,
S.Stinchi,
F.Beltrametti,
A.Lazzarini,
and
S.Donadio
(2003).
The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by nonomuraea species.
|
| |
Chem Biol,
10,
541-549.
|
 |
|
|
|
|
 |
Z.Zhuang,
F.Song,
W.Zhang,
K.Taylor,
A.Archambault,
D.Dunaway-Mariano,
J.Dong,
and
P.R.Carey
(2002).
Kinetic, Raman, NMR, and site-directed mutagenesis studies of the Pseudomonas sp. strain CBS3 4-hydroxybenzoyl-CoA thioesterase active site.
|
| |
Biochemistry,
41,
11152-11160.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2001).
Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies.
|
| |
Annu Rev Biochem,
70,
209-246.
|
 |
|
|
|
|
 |
J.J.Bellizzi,
J.Widom,
C.Kemp,
J.Y.Lu,
A.K.Das,
S.L.Hofmann,
and
J.Clardy
(2000).
The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
|
| |
Proc Natl Acad Sci U S A,
97,
4573-4578.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Devedjiev,
Z.Dauter,
S.R.Kuznetsov,
T.L.Jones,
and
Z.S.Derewenda
(2000).
Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A.
|
| |
Structure,
8,
1137-1146.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

