 |
PDBsum entry 1adn

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transcription regulation
|
PDB id
|
|

|

|
1adn
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.1.1.63
- methylated-DNA--[protein]-cysteine S-methyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a 6-O-methyl-2'-deoxyguanosine in DNA + L-cysteinyl-[protein] = S-methyl-L-cysteinyl-[protein] + a 2'-deoxyguanosine in DNA
|
|
2.
|
a 4-O-methyl-thymidine in DNA + L-cysteinyl-[protein] = a thymidine in DNA + S-methyl-L-cysteinyl-[protein]
|
|
 |
 |
 |
 |
 |
DNA (containing 6-O-methylguanine)
|
+
|
 protein L-cysteine
protein L-cysteine
|
=
|
DNA (without 6-O-methylguanine)
|
+
|
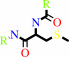 protein S-methyl-L-cysteine
protein S-methyl-L-cysteine
|
|
 |
 |
 |
 |
 |
DNA (containing 4-O-methylthymine)
|
+
|
 protein L-cysteine
protein L-cysteine
|
=
|
DNA (without 4-O-methylthymine)
|
+
|
 protein S-methyl-L-cysteine
protein S-methyl-L-cysteine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.1.1.n11
- ?????
|
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
32:14089-14094
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
Solution structure of the DNA methyl phosphotriester repair domain of Escherichia coli Ada.
|
|
L.C.Myers,
G.L.Verdine,
G.Wagner.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The Escherichia coli Ada protein repairs methyl phosphotriesters in DNA by
direct, irreversible methyl transfer to one of its own cysteine residues. The
methyl-transfer process appears to be autocatalyzed by coordination of the
acceptor residue, Cys-69, to a tightly bound zinc ion. Upon methyl transfer, Ada
acquires the ability to bind DNA sequence-specifically and thereby to induce
genes that confer resistance to methylating agents. The solution structure of an
N-terminal 10-kDa fragment of Ada, which retains zinc binding and DNA methyl
phosphotriester repair activities, was determined using multidimensional
heteronuclear nuclear magnetic resonance techniques. The structure reveals a
zinc-binding motif unlike any observed thus far in transcription factors or
zinc-containing enzymes and provides insight into the mechanism of
metalloactivated DNA repair.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.D.Jones,
R.C.Le Pla,
and
P.B.Farmer
(2010).
Phosphotriester adducts (PTEs): DNA's overlooked lesion.
|
| |
Mutagenesis,
25,
3.
|
 |
|
|
|
|
 |
A.I.Anzellotti,
and
N.P.Farrell
(2008).
Zinc metalloproteins as medicinal targets.
|
| |
Chem Soc Rev,
37,
1629-1651.
|
 |
|
|
|
|
 |
A.M.Krishnakumar,
D.Sliwa,
J.A.Endrizzi,
E.S.Boyd,
S.A.Ensign,
and
J.W.Peters
(2008).
Getting a handle on the role of coenzyme M in alkene metabolism.
|
| |
Microbiol Mol Biol Rev,
72,
445-456.
|
 |
|
|
|
|
 |
G.Parkin
(2007).
Applications of Tripodal [S(3)] and [Se(3)] L(2)X Donor Ligands to Zinc, Cadmium and Mercury Chemistry: Organometallic and Bioinorganic Perspectives.
|
| |
New J Chem,
31,
1996-2014.
|
 |
|
|
|
|
 |
J.Penner-Hahn
(2007).
Zinc-promoted alkyl transfer: a new role for zinc.
|
| |
Curr Opin Chem Biol,
11,
166-171.
|
 |
|
|
|
|
 |
H.Takinowaki,
Y.Matsuda,
T.Yoshida,
Y.Kobayashi,
and
T.Ohkubo
(2006).
The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein.
|
| |
Protein Sci,
15,
487-497.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Mishina,
E.M.Duguid,
and
C.He
(2006).
Direct reversal of DNA alkylation damage.
|
| |
Chem Rev,
106,
215-232.
|
 |
|
|
|
|
 |
W.Maret
(2005).
Zinc coordination environments in proteins determine zinc functions.
|
| |
J Trace Elem Med Biol,
19,
7.
|
 |
|
|
|
|
 |
S.A.Ensign,
and
J.R.Allen
(2003).
Aliphatic epoxide carboxylation.
|
| |
Annu Rev Biochem,
72,
55-76.
|
 |
|
|
|
|
 |
S.S.Krishna,
I.Majumdar,
and
N.V.Grishin
(2003).
Structural classification of zinc fingers: survey and summary.
|
| |
Nucleic Acids Res,
31,
532-550.
|
 |
|
|
|
|
 |
J.G.Krum,
H.Ellsworth,
R.R.Sargeant,
G.Rich,
and
S.A.Ensign
(2002).
Kinetic and microcalorimetric analysis of substrate and cofactor interactions in epoxyalkane:CoM transferase, a zinc-dependent epoxidase.
|
| |
Biochemistry,
41,
5005-5014.
|
 |
|
|
|
|
 |
S.A.Ensign
(2001).
Microbial metabolism of aliphatic alkenes.
|
| |
Biochemistry,
40,
5845-5853.
|
 |
|
|
|
|
 |
Z.J.Sun,
K.S.Kim,
G.Wagner,
and
E.L.Reinherz
(2001).
Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer.
|
| |
Cell,
105,
913-923.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Huang,
K.E.Hightower,
and
C.A.Fierke
(2000).
Mechanistic studies of rat protein farnesyltransferase indicate an associative transition state.
|
| |
Biochemistry,
39,
2593-2602.
|
 |
|
|
|
|
 |
K.Sauer,
and
R.K.Thauer
(2000).
Methyl-coenzyme M formation in methanogenic archaea. Involvement of zinc in coenzyme M activation.
|
| |
Eur J Biochem,
267,
2498-2504.
|
 |
|
|
|
|
 |
K.E.Hightower,
and
C.A.Fierke
(1999).
Zinc-catalyzed sulfur alkyation:insights from protein farnesyltransferase.
|
| |
Curr Opin Chem Biol,
3,
176-181.
|
 |
|
|
|
|
 |
Z.S.Zhou,
K.Peariso,
J.E.Penner-Hahn,
and
R.G.Matthews
(1999).
Identification of the zinc ligands in cobalamin-independent methionine synthase (MetE) from Escherichia coli.
|
| |
Biochemistry,
38,
15915-15926.
|
 |
|
|
|
|
 |
K.E.Hightower,
C.C.Huang,
P.J.Casey,
and
C.A.Fierke
(1998).
H-Ras peptide and protein substrates bind protein farnesyltransferase as an ionized thiolate.
|
| |
Biochemistry,
37,
15555-15562.
|
 |
|
|
|
|
 |
D.L.Pountney,
R.P.Tiwari,
and
J.B.Egan
(1997).
Metal- and DNA-binding properties and mutational analysis of the transcription activating factor, B, of coliphage 186: a prokaryotic C4 zinc-finger protein.
|
| |
Protein Sci,
6,
892-902.
|
 |
|
|
|
|
 |
R.G.Matthews,
and
C.W.Goulding
(1997).
Enzyme-catalyzed methyl transfers to thiols: the role of zinc.
|
| |
Curr Opin Chem Biol,
1,
332-339.
|
 |
|
|
|
|
 |
G.M.LeClerc,
and
D.A.Grahame
(1996).
Methylcobamide:coenzyme M methyltransferase isozymes from Methanosarcina barkeri. Physicochemical characterization, cloning, sequence analysis, and heterologous gene expression.
|
| |
J Biol Chem,
271,
18725-18731.
|
 |
|
|
|
|
 |
J.Habazettl,
L.C.Myers,
F.Yuan,
G.L.Verdine,
and
G.Wagner
(1996).
Backbone dynamics, amide hydrogen exchange, and resonance assignments of the DNA methylphosphotriester repair domain of Escherichia coli Ada using NMR.
|
| |
Biochemistry,
35,
9335-9348.
|
 |
|
|
|
|
 |
J.A.Tainer,
M.M.Thayer,
and
R.P.Cunningham
(1995).
DNA repair proteins.
|
| |
Curr Opin Struct Biol,
5,
20-26.
|
 |
|
|
|
|
 |
L.C.Myers,
F.Jackow,
and
G.L.Verdine
(1995).
Metal dependence of transcriptional switching in Escherichia coli Ada.
|
| |
J Biol Chem,
270,
6664-6670.
|
 |
|
|
|
|
 |
L.H.Pearl,
and
R.Savva
(1995).
DNA repair in three dimensions.
|
| |
Trends Biochem Sci,
20,
421-426.
|
 |
|
|
|
|
 |
G.L.Verdine
(1994).
The flip side of DNA methylation.
|
| |
Cell,
76,
197-200.
|
 |
|
|
|
|
 |
L.C.Myers,
T.D.Cushing,
G.Wagner,
and
G.L.Verdine
(1994).
Metal-coordination sphere in the methylated Ada protein-DNA co-complex.
|
| |
Chem Biol,
1,
91-97.
|
 |
|
|
|
|
 |
L.Szilák,
C.Finta,
A.Patthy,
P.Venetianer,
and
A.Kiss
(1994).
Self-methylation of BspRI DNA-methyltransferase.
|
| |
Nucleic Acids Res,
22,
2876-2881.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

