 |
PDBsum entry 1a4x

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transcription regulation
|
PDB id
|
|

|

|
1a4x
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.9
- uracil phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
UMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + uracil
|
 |
 |
 |
 |
 |
 UMP
UMP
|
+
|
 diphosphate
diphosphate
|
=
|
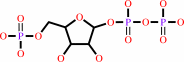 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
+
|
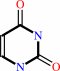 uracil
uracil
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
6:337-350
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
|
|
D.R.Tomchick,
R.J.Turner,
R.L.Switzer,
J.L.Smith.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: The expression of pyrimidine nucleotide biosynthetic (pyr) genes in
Bacillus subtilis is regulated by transcriptional attenuation. The PyrR
attenuation protein binds to specific sites in pyr mRNA, allowing the formation
of downstream terminator structures. UMP and 5-phosphoribosyl-1-pyrophosphate
(PRPP), a nucleotide metabolite, are co-regulators with PyrR. The smallest RNA
shown to bind tightly to PyrR is a 28-30 nucleotide stem-loop that contains a
purine-rich bulge and a putative-GNRA tetraloop. PyrR is also a uracil
phosphoribosyltransferase (UPRTase), although the relationship between enzymatic
activity and RNA recognition is unclear, and the UPRTase activity of PyrR is not
physiologically significant in B. subtilis. Elucidating the role of PyrR
structural motifs in UMP-dependent RNA binding is an important step towards
understanding the mechanism of pyr transcriptional attenuation. RESULTS: The 1.6
A crystal structure of B. subtilis PyrR has been determined by multiwavelength
anomalous diffraction, using a Sm co-crystal. As expected, the structure of PyrR
is homologous to those proteins of the large type I PRTase structural family; it
is most similar to hypoxanthine-guanine-xanthine PRTase (HGXPRTase). The PyrR
dimer differs from other PRTase dimers, suggesting it may have evolved
specifically for RNA binding. A large, basic, surface at the dimer interface is
an obvious RNA-binding site and uracil specificity is probably provided by
hydrogen bonds from mainchain and sidechain atoms in the hood subdomain. These
models of RNA and UMP binding are consistent with biological data. CONCLUSIONS:
The B. subtilis protein PyrR has adapted the substrate- and product-binding
capacities of a PRTase, probably an HGXPRTase, producing a new regulatory
function in which the substrate and product are co-regulators of transcription
termination. The structure is consistent with the idea that PyrR regulatory
function is independent of catalytic activity, which is likely to be extremely
low under physiological conditions.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 6.
Figure 6. Stereo view of a model of UMP bound to the active
site of the PyrR monomer from the dimer crystal structure.
Protein residues implicated in catalytic activity and/or UMP
coordination are shown with black bonds; UMP is shown with white
bonds; carbon atoms are white; oxygen, nitrogen and phosphorus
are shaded. Potential hydrogen bonds between the UMP base and
protein are shown as dashed lines. One of these hydrogen bonds
is to invariant Arg138 in the dimer loop. The figure was
prepared with the program MOLSCRIPT [71].
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1998,
6,
337-350)
copyright 1998.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
R.Kumar,
P.Shah,
E.Swiatlo,
S.C.Burgess,
M.L.Lawrence,
and
B.Nanduri
(2010).
Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays.
|
| |
BMC Genomics,
11,
350.
|
 |
|
|
|
|
 |
S.E.Mainguet,
B.Gakière,
A.Majira,
S.Pelletier,
F.Bringel,
F.Guérard,
M.Caboche,
R.Berthomé,
and
J.P.Renou
(2009).
Uracil salvage is necessary for early Arabidopsis development.
|
| |
Plant J,
60,
280-291.
|
 |
|
|
|
|
 |
C.L.Turnbough,
and
R.L.Switzer
(2008).
Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors.
|
| |
Microbiol Mol Biol Rev,
72,
266.
|
 |
|
|
|
|
 |
C.M.Jørgensen,
C.J.Fields,
P.Chander,
D.Watt,
J.W.Burgner,
J.L.Smith,
and
R.L.Switzer
(2008).
pyr RNA binding to the Bacillus caldolyticus PyrR attenuation protein - characterization and regulation by uridine and guanosine nucleotides.
|
| |
FEBS J,
275,
655-670.
|
 |
|
|
|
|
 |
H.Tan,
S.Wu,
J.Wang,
and
Z.K.Zhao
(2008).
The JMJD2 members of histone demethylase revisited.
|
| |
Mol Biol Rep,
35,
551-556.
|
 |
|
|
|
|
 |
R.Arreola,
A.Vega-Miranda,
A.Gómez-Puyou,
R.Pérez-Montfort,
E.Merino-Pérez,
and
A.Torres-Larios
(2008).
Expression, purification and preliminary X-ray diffraction studies of the transcriptional factor PyrR from Bacillus halodurans.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
692-696.
|
 |
|
|
|
|
 |
F.Arsène-Ploetze,
H.Nicoloff,
B.Kammerer,
J.Martinussen,
and
F.Bringel
(2006).
Uracil salvage pathway in Lactobacillus plantarum: Transcription and genetic studies.
|
| |
J Bacteriol,
188,
4777-4786.
|
 |
|
|
|
|
 |
F.Arsène-Ploetze,
V.Kugler,
J.Martinussen,
and
F.Bringel
(2006).
Expression of the pyr operon of Lactobacillus plantarum is regulated by inorganic carbon availability through a second regulator, PyrR2, homologous to the pyrimidine-dependent regulator PyrR1.
|
| |
J Bacteriol,
188,
8607-8616.
|
 |
|
|
|
|
 |
Z.Chen,
J.Zang,
J.Whetstine,
X.Hong,
F.Davrazou,
T.G.Kutateladze,
M.Simpson,
Q.Mao,
C.H.Pan,
S.Dai,
J.Hagman,
K.Hansen,
Y.Shi,
and
G.Zhang
(2006).
Structural insights into histone demethylation by JMJD2 family members.
|
| |
Cell,
125,
691-702.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Janner
(2005).
Strongly correlated structure of axial-symmetric proteins. I. Orthorhombic, tetragonal, trigonal and hexagonal symmetries.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
247-255.
|
 |
|
|
|
|
 |
H.Nicoloff,
A.Elagöz,
F.Arsène-Ploetze,
B.Kammerer,
J.Martinussen,
and
F.Bringel
(2005).
Repression of the pyr operon in Lactobacillus plantarum prevents its ability to grow at low carbon dioxide levels.
|
| |
J Bacteriol,
187,
2093-2104.
|
 |
|
|
|
|
 |
K.A.Kantardjieff,
C.Vasquez,
P.Castro,
N.M.Warfel,
B.S.Rho,
T.Lekin,
C.Y.Kim,
B.W.Segelke,
T.C.Terwilliger,
and
B.Rupp
(2005).
Structure of pyrR (Rv1379) from Mycobacterium tuberculosis: a persistence gene and protein drug target.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
355-364.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Chander,
K.M.Halbig,
J.K.Miller,
C.J.Fields,
H.K.Bonner,
G.K.Grabner,
R.L.Switzer,
and
J.L.Smith
(2005).
Structure of the nucleotide complex of PyrR, the pyr attenuation protein from Bacillus caldolyticus, suggests dual regulation by pyrimidine and purine nucleotides.
|
| |
J Bacteriol,
187,
1773-1782.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.K.Grabner,
and
R.L.Switzer
(2003).
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine attenuation regulatory protein PyrR.
|
| |
J Biol Chem,
278,
6921-6927.
|
 |
|
|
|
|
 |
S.C.Sinha,
J.Krahn,
B.S.Shin,
D.R.Tomchick,
H.Zalkin,
and
J.L.Smith
(2003).
The purine repressor of Bacillus subtilis: a novel combination of domains adapted for transcription regulation.
|
| |
J Bacteriol,
185,
4087-4098.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.K.Savacool,
and
R.L.Switzer
(2002).
Characterization of the interaction of Bacillus subtilis PyrR with pyr mRNA by site-directed mutagenesis of the protein.
|
| |
J Bacteriol,
184,
2521-2528.
|
 |
|
|
|
|
 |
J.C.Evans,
D.P.Huddler,
J.Jiracek,
C.Castro,
N.S.Millian,
T.A.Garrow,
and
M.L.Ludwig
(2002).
Betaine-homocysteine methyltransferase: zinc in a distorted barrel.
|
| |
Structure,
10,
1159-1171.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.A.Schumacher,
C.J.Bashor,
M.H.Song,
K.Otsu,
S.Zhu,
R.J.Parry,
B.Ullman,
and
R.G.Brennan
(2002).
The structural mechanism of GTP stabilized oligomerization and catalytic activation of the Toxoplasma gondii uracil phosphoribosyltransferase.
|
| |
Proc Natl Acad Sci U S A,
99,
78-83.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.R.Bonner,
J.N.D'Elia,
B.K.Billips,
and
R.L.Switzer
(2001).
Molecular recognition of pyr mRNA by the Bacillus subtilis attenuation regulatory protein PyrR.
|
| |
Nucleic Acids Res,
29,
4851-4865.
|
 |
|
|
|
|
 |
J.Martinussen,
J.Schallert,
B.Andersen,
and
K.Hammer
(2001).
The pyrimidine operon pyrRPB-carA from Lactococcus lactis.
|
| |
J Bacteriol,
183,
2785-2794.
|
 |
|
|
|
|
 |
C.Yanofsky
(2000).
Transcription attenuation: once viewed as a novel regulatory strategy.
|
| |
J Bacteriol,
182,
1-8.
|
 |
|
|
|
|
 |
M.Oda,
N.Kobayashi,
A.Ito,
Y.Kurusu,
and
K.Taira
(2000).
cis-acting regulatory sequences for antitermination in the transcript of the Bacillus subtilis hut operon and histidine-dependent binding of HutP to the transcript containing the regulatory sequences.
|
| |
Mol Microbiol,
35,
1244-1254.
|
 |
|
|
|
|
 |
T.M.Henkin
(2000).
Transcription termination control in bacteria.
|
| |
Curr Opin Microbiol,
3,
149-153.
|
 |
|
|
|
|
 |
S.Cusack
(1999).
RNA-protein complexes.
|
| |
Curr Opin Struct Biol,
9,
66-73.
|
 |
|
|
|
|
 |
A.W.Tsang,
and
J.C.Escalante-Semerena
(1998).
CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2.
|
| |
J Biol Chem,
273,
31788-31794.
|
 |
|
|
|
|
 |
J.L.Smith
(1998).
Glutamine PRPP amidotransferase: snapshots of an enzyme in action.
|
| |
Curr Opin Struct Biol,
8,
686-694.
|
 |
|
|
|
|
 |
V.Sharma,
C.Grubmeyer,
and
J.C.Sacchettini
(1998).
Crystal structure of quinolinic acid phosphoribosyltransferase from Mmycobacterium tuberculosis: a potential TB drug target.
|
| |
Structure,
6,
1587-1599.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

