 |
PDBsum entry 1a05

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1a05
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of the complex of 3-isopropylmalate dehydrogenase from thiobacillus ferrooxidans with 3-isopropylmalate
|
|
Structure:
|
 |
3-isopropylmalate dehydrogenase. Chain: a, b. Synonym: ipmdh, imdh. Engineered: yes
|
|
Source:
|
 |
Acidithiobacillus ferrooxidans. Organism_taxid: 920. Strain: ap19-3. Gene: leub. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.198
|
R-free:
|
0.275
|
|
|
Authors:
|
 |
K.Imada,K.Inagaki,H.Matsunami,H.Kawaguchi,H.Tanaka,N.Tanaka,K.Namba
|
Key ref:

|
 |
K.Imada
et al.
(1998).
Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure,
6,
971-982.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
09-Dec-97
|
Release date:
|
17-Jun-98
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q56268
(LEU3_ACIFR) -
3-isopropylmalate dehydrogenase from Acidithiobacillus ferrooxidans
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
358 a.a.
357 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.85
- 3-isopropylmalate dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3S)-3-isopropylmalate + NAD+ = 4-methyl-2-oxopentanoate + CO2 + NADH
|
 |
 |
 |
 |
 |
(2R,3S)-3-isopropylmalate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NAD(+)
NAD(+)
|
=
|
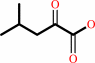 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
 CO2
CO2
|
+
|
 NADH
NADH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
6:971-982
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
|
|
K.Imada,
K.Inagaki,
H.Matsunami,
H.Kawaguchi,
H.Tanaka,
N.Tanaka,
K.Namba.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: 3-Isopropylmalate dehydrogenase (IPMDH) and isocitrate dehydrogenase
(ICDH) belong to a unique family of bifunctional decarboxylating dehydrogenases.
Although the ICDH dimer catalyzes its reaction under a closed conformation,
known structures of the IPMDH dimer (without substrate) adopt a fully open or a
partially closed form. Considering the similarity in the catalytic mechanism,
the IPMDH dimer must be in a fully closed conformation during the reaction. A
large conformational change should therefore occur upon substrate binding.
RESULTS: We have determined the crystal structure of IPMDH from Thiobacillus
ferrooxidans (Tf) complexed with 3-isopropylmalate (IPM) at 2.0 A resolution by
the molecular replacement method. The structure shows a fully closed
conformation and the substrate-binding site is quite similar to that of ICDH
except for a region around the gamma-isopropyl group. The gamma group is
recognized by a unique hydrophobic pocket, which includes Glu88, Leu91 and Leu92
from subunit 1 and Val193' from subunit 2. CONCLUSIONS: A large movement of
domain 1 is induced by substrate binding, which results in the formation of the
hydrophobic pocket for the gamma-isopropyl moiety of IPM. A glutamic acid in
domain 1, Glu88, participates in the formation of the hydrophobic pocket. The C
beta and C gamma atoms of Glu88 interact with the gamma-isopropyl moiety of IPM
and are central to the recognition of substrate. The acidic tip of Glu88 is
likely to interact with the nicotinamide mononucleotide (NMN) ribose of NAD+ in
the ternary complex. This structure clearly explains the substrate specificity
of IPMDH.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 6.
Figure 6. Comparison of the g moiety recognition site. (a)
A space-filling stereo drawing of the active site of Tf-IPMDH,
viewed from the right-hand side of Figure 7b. The substrate is
shown in ball-and-stick representation. Atoms are color-coded:
oxygen, red; nitrogen, blue; and carbon, gray. A green ball
represents a sulfur atom of a methionine residue and a small
magenta ball indicates the magnesium ion. The carbon atoms in
the hydrophobic pocket are highlighted in orange. Residues
forming the hydrophobic pocket are labeled with their residue
numbers. Schematic representations of the active site of (b)
Tf-IPMDH (closed conformation), (c) Ec-ICDH (closed
conformation), (d) St-IPMDH (closed conformation), (e) Tt-IPMDH
IPM complex (open conformation). The orientation is as in (a).
(d) Was generated by the superposition of IPM and Mg of the
Tf-IPMDH binary complex onto the structure of St-IPMDH solved
without IPM and Mg. The protein backbone is shown in a ribbon
representation. The substrate molecules and sidechains lying in
the active sites are shown in ball-and-stick representation. The
color coding of the atoms is as in (a). Residues interacting
with the g moiety of the substrates are labeled with their
residue numbers. The figures were generated with MOLSCRIPT [36]
and RASTER3D [37].
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1998,
6,
971-982)
copyright 1998.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
T.Nagae,
T.Kawamura,
L.M.Chavas,
K.Niwa,
M.Hasegawa,
C.Kato,
and
N.Watanabe
(2012).
High-pressure-induced water penetration into 3-isopropylmalate dehydrogenase.
|
| |
Acta Crystallogr D Biol Crystallogr,
68,
300-309.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Ã.‰.Gráczer,
A.Merli,
R.K.Singh,
M.Karuppasamy,
P.Závodszky,
M.S.Weiss,
and
M.Vas
(2011).
Atomic level description of the domain closure in a dimeric enzyme: thermus thermophilus 3-isopropylmalate dehydrogenase.
|
| |
Mol Biosyst,
7,
1646-1659.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Lunzer,
G.B.Golding,
and
A.M.Dean
(2010).
Pervasive cryptic epistasis in molecular evolution.
|
| |
PLoS Genet,
6,
e1001162.
|
 |
|
|
|
|
 |
M.Röttig,
C.Rausch,
and
O.Kohlbacher
(2010).
Combining structure and sequence information allows automated prediction of substrate specificities within enzyme families.
|
| |
PLoS Comput Biol,
6,
e1000636.
|
 |
|
|
|
|
 |
I.Hajdú,
A.Szilágyi,
J.Kardos,
and
P.Závodszky
(2009).
A link between hinge-bending domain motions and the temperature dependence of catalysis in 3-isopropylmalate dehydrogenase.
|
| |
Biophys J,
96,
5003-5012.
|
 |
|
|
|
|
 |
J.R.Weber
(2009).
ProteinShader: illustrative rendering of macromolecules.
|
| |
BMC Struct Biol,
9,
19.
|
 |
|
|
|
|
 |
R.Kasahara,
T.Sato,
H.Tamegai,
and
C.Kato
(2009).
Piezo-adapted 3-isopropylmalate dehydrogenase of the obligate piezophile Shewanella benthica DB21MT-2 isolated from the 11,000-m depth of the Mariana Trench.
|
| |
Biosci Biotechnol Biochem,
73,
2541-2543.
|
 |
|
|
|
|
 |
K.Imada,
T.Tamura,
R.Takenaka,
I.Kobayashi,
K.Namba,
and
K.Inagaki
(2008).
Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity.
|
| |
Proteins,
70,
63-71.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Sasaki,
M.Uno,
S.Akanuma,
and
A.Yamagishi
(2008).
Random mutagenesis improves the low-temperature activity of the tetrameric 3-isopropylmalate dehydrogenase from the hyperthermophile Sulfolobus tokodaii.
|
| |
Protein Eng Des Sel,
21,
721-727.
|
 |
|
|
|
|
 |
R.Stokke,
D.Madern,
A.E.Fedøy,
S.Karlsen,
N.K.Birkeland,
and
I.H.Steen
(2007).
Biochemical characterization of isocitrate dehydrogenase from Methylococcus capsulatus reveals a unique NAD+-dependent homotetrameric enzyme.
|
| |
Arch Microbiol,
187,
361-370.
|
 |
|
|
|
|
 |
J.A.McCourt,
and
R.G.Duggleby
(2006).
Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids.
|
| |
Amino Acids,
31,
173-210.
|
 |
|
|
|
|
 |
O.V.Kalinina,
and
M.S.Gelfand
(2006).
Amino acid residues that determine functional specificity of NADP- and NAD-dependent isocitrate and isopropylmalate dehydrogenases.
|
| |
Proteins,
64,
1001-1009.
|
 |
|
|
|
|
 |
H.Turakainen,
and
M.Korhola
(2005).
Cloning, sequencing and application of the LEU2 gene from the sour dough yeast Candida milleri.
|
| |
Yeast,
22,
805-812.
|
 |
|
|
|
|
 |
J.Miyazaki,
K.Asada,
S.Fushinobu,
T.Kuzuyama,
and
M.Nishiyama
(2005).
Crystal structure of tetrameric homoisocitrate dehydrogenase from an extreme thermophile, Thermus thermophilus: involvement of hydrophobic dimer-dimer interaction in extremely high thermotolerance.
|
| |
J Bacteriol,
187,
6779-6788.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Dupuis,
J.F.Sadoc,
and
J.P.Mornon
(2004).
Protein secondary structure assignment through Voronoï tessellation.
|
| |
Proteins,
55,
519-528.
|
 |
|
|
|
|
 |
J.Miyazaki,
N.Kobashi,
M.Nishiyama,
and
H.Yamane
(2003).
Characterization of homoisocitrate dehydrogenase involved in lysine biosynthesis of an extremely thermophilic bacterium, Thermus thermophilus HB27, and evolutionary implication of beta-decarboxylating dehydrogenase.
|
| |
J Biol Chem,
278,
1864-1871.
|
 |
|
|
|
|
 |
J.Sivaraman,
Y.Li,
J.Banks,
D.E.Cane,
A.Matte,
and
M.Cygler
(2003).
Crystal structure of Escherichia coli PdxA, an enzyme involved in the pyridoxal phosphate biosynthesis pathway.
|
| |
J Biol Chem,
278,
43682-43690.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Yasutake,
S.Watanabe,
M.Yao,
Y.Takada,
N.Fukunaga,
and
I.Tanaka
(2003).
Crystal structure of the monomeric isocitrate dehydrogenase in the presence of NADP+: insight into the cofactor recognition, catalysis, and evolution.
|
| |
J Biol Chem,
278,
36897-36904.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Inoue,
T.Tamura,
N.Ehara,
A.Nishito,
Y.Nakayama,
M.Maekawa,
K.Imada,
H.Tanaka,
and
K.Inagaki
(2002).
Biochemical and molecular characterization of the NAD(+)-dependent isocitrate dehydrogenase from the chemolithotroph Acidithiobacillus thiooxidans.
|
| |
FEMS Microbiol Lett,
214,
127-132.
|
 |
|
|
|
|
 |
M.Karlström,
I.H.Steen,
G.Tibbelin,
T.Lien,
N.K.Birkeland,
and
R.Ladenstein
(2002).
Crystallization and preliminary X-ray structure analysis of isocitrate dehydrogenase from two hyperthermophiles, Aeropyrum pernix and Thermotoga maritima.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
2162-2164.
|
 |
|
|
|
|
 |
M.Fujita,
H.Tamegai,
T.Eguchi,
and
K.Kakinuma
(2001).
Novel substrate specificity of designer 3-isopropylmalate dehydrogenase derived from Thermus thermophilus HB8.
|
| |
Biosci Biotechnol Biochem,
65,
2695-2700.
|
 |
|
|
|
|
 |
S.A.Doyle,
P.T.Beernink,
and
D.E.Koshland
(2001).
Structural basis for a change in substrate specificity: crystal structure of S113E isocitrate dehydrogenase in a complex with isopropylmalate, Mg2+, and NADP.
|
| |
Biochemistry,
40,
4234-4241.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Kawaguchi,
K.Inagaki,
H.Matsunami,
Y.Nakayama,
T.Tano,
and
H.Tanaka
(2000).
Purification and characterization of 3-isopropylmalate dehydrogenase from Thiobacillus thiooxidans.
|
| |
J Biosci Bioeng,
90,
459-461.
|
 |
|
|
|
|
 |
S.A.Doyle,
S.Y.Fung,
and
D.E.Koshland
(2000).
Redesigning the substrate specificity of an enzyme: isocitrate dehydrogenase.
|
| |
Biochemistry,
39,
14348-14355.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

