 |
PDBsum entry 1q2u

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
RNA binding protein
|
PDB id
|
|

|

|
1q2u
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.2.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.5.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.5.1.124
- protein deglycase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N(omega)-(1-hydroxy-2-oxopropyl)-L-arginyl-[protein] + H2O = lactate + L-arginyl-[protein] + H+
|
|
2.
|
N6-(1-hydroxy-2-oxopropyl)-L-lysyl-[protein] + H2O = lactate + L-lysyl-[protein] + H+
|
|
3.
|
S-(1-hydroxy-2-oxopropyl)-L-cysteinyl-[protein] + H2O = lactate + L-cysteinyl-[protein] + H+
|
|
 |
 |
 |
 |
 |
N(omega)-(1-hydroxy-2-oxopropyl)-L-arginyl-[protein]
|
+
|
H2O
|
=
|
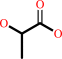 lactate
lactate
|
+
|
L-arginyl-[protein]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
N(6)-(1-hydroxy-2-oxopropyl)-L-lysyl-[protein]
|
+
|
H2O
|
=
|
 lactate
lactate
|
+
|
L-lysyl-[protein]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
S-(1-hydroxy-2-oxopropyl)-L-cysteinyl-[protein]
|
+
|
H2O
|
=
|
 lactate
lactate
|
+
|
L-cysteinyl-[protein]
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
FEBS Lett
549:171-175
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of DJ-1/RS and implication on familial Parkinson's disease.
|
|
Q.Huai,
Y.Sun,
H.Wang,
L.S.Chin,
L.Li,
H.Robinson,
H.Ke.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
DJ-1 is a protein involved in multiple physiological processes, including
cancer, Parkinson's disease, and male fertility. It is unknown how DJ-1
functions in the apparently different systems. The crystal structure of DJ-1 at
1.6 A resolution shows that DJ-1 is a helix-strand-helix sandwich and forms a
dimer. The DJ-1 structure is similar to the members of the intracellular
protease PfpI family. However, the catalytic triad of Cys-His-Glu is not
strictly conserved in DJ-1, implying that DJ-1 has a different catalytic
mechanism if it acts as a protease or DJ-1 serves as a regulatory protein in the
physiological processes. The structure shows that Leu166 positions in the middle
of a helix and thus predicts that the L166P mutation will bend the helix and
impact the dimerization of DJ-1. As a result, the conformational changes may
diminish the DJ-1 binding with its partner, leading to the familial Parkinson's
disease caused by the single L166P mutation.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |

|
 |

|
 |
Figure 1.
Fig. 1. Ribbon representation of monomeric DJ-1 in two
different views (a and b), and DJ-1 dimer (c and d). The red
balls in c and d represent Leu166 that is located in the middle
of helix H7. The purple balls are Lys130, (e) the secondary
structure and sequence.
|
 |
Figure 2.
Fig. 2. Superposition of DJ-1 (golden) over intracellular
protease PH1704 (cyan, left) and E. coli heat shock protein
HSP31 (green, right).
|
 |
Figure 3.
Fig. 3. The catalytic triad of Cys100–His101 and Glu74 at
the active site of PH1704 (left). The dotted lines represent the
hydrogen bonds. Residue Glu74 comes from the neighboring subunit
in the hexamer of PH1704. Right, a putative binding pocket of
DJ-1. The catalytic triad is not conserved in DJ-1, but the
pocket may still be capable of binding with its substrate
proteins.
|
 |
Figure 4.
Fig. 4. The interfacial region of DJ-1 dimer and the
position of Leu166. Two helices H7 and H8 are shown in the
similar orientation as in Fig. 1d. The L166P mutation is
expected to cause significant conformational changes and to
impact the dimerization of DJ-1.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Federation of European Biochemical Societies:
FEBS Lett
(2003,
549,
171-175)
copyright 2003.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.P.Ramsey,
and
B.I.Giasson
(2010).
L10p and P158DEL DJ-1 mutations cause protein instability, aggregation, and dimerization impairments.
|
| |
J Neurosci Res,
88,
3111-3124.
|
 |
|
|
|
|
 |
C.P.Ramsey,
E.Tsika,
H.Ischiropoulos,
and
B.I.Giasson
(2010).
DJ-1 deficient mice demonstrate similar vulnerability to pathogenic Ala53Thr human alpha-syn toxicity.
|
| |
Hum Mol Genet,
19,
1425-1437.
|
 |
|
|
|
|
 |
J.Chen,
L.Li,
and
L.S.Chin
(2010).
Parkinson disease protein DJ-1 converts from a zymogen to a protease by carboxyl-terminal cleavage.
|
| |
Hum Mol Genet,
19,
2395-2408.
|
 |
|
|
|
|
 |
L.S.Chin,
J.A.Olzmann,
and
L.Li
(2010).
Parkin-mediated ubiquitin signalling in aggresome formation and autophagy.
|
| |
Biochem Soc Trans,
38,
144-149.
|
 |
|
|
|
|
 |
E.Junn,
W.H.Jang,
X.Zhao,
B.S.Jeong,
and
M.M.Mouradian
(2009).
Mitochondrial localization of DJ-1 leads to enhanced neuroprotection.
|
| |
J Neurosci Res,
87,
123-129.
|
 |
|
|
|
|
 |
K.Yamane,
Y.Kitamura,
T.Yanagida,
K.Takata,
D.Yanagisawa,
T.Taniguchi,
T.Taira,
and
H.Ariga
(2009).
Oxidative Neurodegeneration Is Prevented by UCP0045037, an Allosteric Modulator for the Reduced Form of DJ-1, a Wild-Type of Familial Parkinson's Disease-Linked PARK7.
|
| |
Int J Mol Sci,
10,
4789-4804.
|
 |
|
|
|
|
 |
P.J.Kahle,
J.Waak,
and
T.Gasser
(2009).
DJ-1 and prevention of oxidative stress in Parkinson's disease and other age-related disorders.
|
| |
Free Radic Biol Med,
47,
1354-1361.
|
 |
|
|
|
|
 |
R.J.Keyser,
L.van der Merwe,
M.Venter,
C.Kinnear,
L.Warnich,
J.Carr,
and
S.Bardien
(2009).
Identification of a novel functional deletion variant in the 5'-UTR of the DJ-1 gene.
|
| |
BMC Med Genet,
10,
105.
|
 |
|
|
|
|
 |
T.Yanagida,
J.Tsushima,
Y.Kitamura,
D.Yanagisawa,
K.Takata,
T.Shibaike,
A.Yamamoto,
T.Taniguchi,
H.Yasui,
T.Taira,
S.Morikawa,
T.Inubushi,
I.Tooyama,
and
H.Ariga
(2009).
Oxidative stress induction of DJ-1 protein in reactive astrocytes scavenges free radicals and reduces cell injury.
|
| |
Oxid Med Cell Longev,
2,
36-42.
|
 |
|
|
|
|
 |
A.C.Witt,
M.Lakshminarasimhan,
B.C.Remington,
S.Hasim,
E.Pozharski,
and
M.A.Wilson
(2008).
Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
|
| |
Biochemistry,
47,
7430-7440.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.P.Ramsey,
and
B.I.Giasson
(2008).
The E163K DJ-1 mutant shows specific antioxidant deficiency.
|
| |
Brain Res,
1239,
1.
|
 |
|
|
|
|
 |
D.Yanagisawa,
Y.Kitamura,
M.Inden,
K.Takata,
T.Taniguchi,
S.Morikawa,
M.Morita,
T.Inubushi,
I.Tooyama,
T.Taira,
S.M.Iguchi-Ariga,
A.Akaike,
and
H.Ariga
(2008).
DJ-1 protects against neurodegeneration caused by focal cerebral ischemia and reperfusion in rats.
|
| |
J Cereb Blood Flow Metab,
28,
563-578.
|
 |
|
|
|
|
 |
G.Malgieri,
and
D.Eliezer
(2008).
Structural effects of Parkinson's disease linked DJ-1 mutations.
|
| |
Protein Sci,
17,
855-868.
|
 |
|
|
|
|
 |
M.Lakshminarasimhan,
M.T.Maldonado,
W.Zhou,
A.L.Fink,
and
M.A.Wilson
(2008).
Structural impact of three Parkinsonism-associated missense mutations on human DJ-1.
|
| |
Biochemistry,
47,
1381-1392.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.S.Cha,
H.I.Jung,
H.Jeon,
Y.J.An,
I.K.Kim,
S.Yun,
H.J.Ahn,
K.C.Chung,
S.H.Lee,
P.G.Suh,
and
S.O.Kang
(2008).
Crystal Structure of Filamentous Aggregates of Human DJ-1 Formed in an Inorganic Phosphate-dependent Manner.
|
| |
J Biol Chem,
283,
34069-34075.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.A.Khalil,
and
P.James
(2007).
Biomarker discovery: a proteomic approach for brain cancer profiling.
|
| |
Cancer Sci,
98,
201-213.
|
 |
|
|
|
|
 |
J.A.Olzmann,
J.R.Bordelon,
E.C.Muly,
H.D.Rees,
A.I.Levey,
L.Li,
and
L.S.Chin
(2007).
Selective enrichment of DJ-1 protein in primate striatal neuronal processes: implications for Parkinson's disease.
|
| |
J Comp Neurol,
500,
585-599.
|
 |
|
|
|
|
 |
J.L.Jiménez,
B.Hegemann,
J.R.Hutchins,
J.M.Peters,
and
R.Durbin
(2007).
A systematic comparative and structural analysis of protein phosphorylation sites based on the mtcPTM database.
|
| |
Genome Biol,
8,
R90.
|
 |
|
|
|
|
 |
K.Görner,
E.Holtorf,
J.Waak,
T.T.Pham,
D.M.Vogt-Weisenhorn,
W.Wurst,
C.Haass,
and
P.J.Kahle
(2007).
Structural determinants of the C-terminal helix-kink-helix motif essential for protein stability and survival promoting activity of DJ-1.
|
| |
J Biol Chem,
282,
13680-13691.
|
 |
|
|
|
|
 |
N.Inamdar,
D.Arulmozhi,
A.Tandon,
and
S.Bodhankar
(2007).
Parkinson's disease: genetics and beyond.
|
| |
Curr Neuropharmacol,
5,
99.
|
 |
|
|
|
|
 |
J.Choi,
M.C.Sullards,
J.A.Olzmann,
H.D.Rees,
S.T.Weintraub,
D.E.Bostwick,
M.Gearing,
A.I.Levey,
L.S.Chin,
and
L.Li
(2006).
Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases.
|
| |
J Biol Chem,
281,
10816-10824.
|
 |
|
|
|
|
 |
M.C.Meulener,
K.Xu,
L.Thomson,
L.Thompson,
H.Ischiropoulos,
and
N.M.Bonini
(2006).
Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging.
|
| |
Proc Natl Acad Sci U S A,
103,
12517-12522.
|
 |
|
|
|
|
 |
M.Mujacic,
and
F.Baneyx
(2006).
Regulation of Escherichia coli hchA, a stress-inducible gene encoding molecular chaperone Hsp31.
|
| |
Mol Microbiol,
60,
1576-1589.
|
 |
|
|
|
|
 |
S.Kubo,
N.Hattori,
and
Y.Mizuno
(2006).
Recessive Parkinson's disease.
|
| |
Mov Disord,
21,
885-893.
|
 |
|
|
|
|
 |
Y.Shinbo,
T.Niki,
T.Taira,
H.Ooe,
K.Takahashi-Niki,
C.Maita,
C.Seino,
S.M.Iguchi-Ariga,
and
H.Ariga
(2006).
Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities.
|
| |
Cell Death Differ,
13,
96.
|
 |
|
|
|
|
 |
L.Chen,
B.Cagniard,
T.Mathews,
S.Jones,
H.C.Koh,
Y.Ding,
P.M.Carvey,
Z.Ling,
U.J.Kang,
and
X.Zhuang
(2005).
Age-dependent motor deficits and dopaminergic dysfunction in DJ-1 null mice.
|
| |
J Biol Chem,
280,
21418-21426.
|
 |
|
|
|
|
 |
R.H.Kim,
M.Peters,
Y.Jang,
W.Shi,
M.Pintilie,
G.C.Fletcher,
C.DeLuca,
J.Liepa,
L.Zhou,
B.Snow,
R.C.Binari,
A.S.Manoukian,
M.R.Bray,
F.F.Liu,
M.S.Tsao,
and
T.W.Mak
(2005).
DJ-1, a novel regulator of the tumor suppressor PTEN.
|
| |
Cancer Cell,
7,
263-273.
|
 |
|
|
|
|
 |
J.A.Olzmann,
K.Brown,
K.D.Wilkinson,
H.D.Rees,
Q.Huai,
H.Ke,
A.I.Levey,
L.Li,
and
L.S.Chin
(2004).
Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function.
|
| |
J Biol Chem,
279,
8506-8515.
|
 |
|
|
|
|
 |
K.Görner,
E.Holtorf,
S.Odoy,
B.Nuscher,
A.Yamamoto,
J.T.Regula,
K.Beyer,
C.Haass,
and
P.J.Kahle
(2004).
Differential effects of Parkinson's disease-associated mutations on stability and folding of DJ-1.
|
| |
J Biol Chem,
279,
6943-6951.
|
 |
|
|
|
|
 |
M.A.Wilson,
C.V.St Amour,
J.L.Collins,
D.Ringe,
and
G.A.Petsko
(2004).
The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: a member of the DJ-1/ThiJ/PfpI superfamily.
|
| |
Proc Natl Acad Sci U S A,
101,
1531-1536.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Graille,
S.Quevillon-Cheruel,
N.Leulliot,
C.Z.Zhou,
I.Li de la Sierra Gallay,
L.Jacquamet,
J.L.Ferrer,
D.Liger,
A.Poupon,
J.Janin,
and
H.van Tilbeurgh
(2004).
Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family.
|
| |
Structure,
12,
839-847.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Mujacic,
M.W.Bader,
and
F.Baneyx
(2004).
Escherichia coli Hsp31 functions as a holding chaperone that cooperates with the DnaK-DnaJ-GrpE system in the management of protein misfolding under severe stress conditions.
|
| |
Mol Microbiol,
51,
849-859.
|
 |
|
|
|
|
 |
M.S.Sastry,
P.M.Quigley,
W.G.Hol,
and
F.Baneyx
(2004).
The linker-loop region of Escherichia coli chaperone Hsp31 functions as a gate that modulates high-affinity substrate binding at elevated temperatures.
|
| |
Proc Natl Acad Sci U S A,
101,
8587-8592.
|
 |
|
|
|
|
 |
R.M.Canet-Avilés,
M.A.Wilson,
D.W.Miller,
R.Ahmad,
C.McLendon,
S.Bandyopadhyay,
M.J.Baptista,
D.Ringe,
G.A.Petsko,
and
M.R.Cookson
(2004).
The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization.
|
| |
Proc Natl Acad Sci U S A,
101,
9103-9108.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Bandyopadhyay,
and
M.R.Cookson
(2004).
Evolutionary and functional relationships within the DJ1 superfamily.
|
| |
BMC Evol Biol,
4,
6.
|
 |
|
|
|
|
 |
S.Shendelman,
A.Jonason,
C.Martinat,
T.Leete,
and
A.Abeliovich
(2004).
DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation.
|
| |
PLoS Biol,
2,
e362.
|
 |
|
|
|
|
 |
T.Taira,
Y.Saito,
T.Niki,
S.M.Iguchi-Ariga,
K.Takahashi,
and
H.Ariga
(2004).
DJ-1 has a role in antioxidative stress to prevent cell death.
|
| |
EMBO Rep,
5,
213-218.
|
 |
|
|
|
|
 |
V.A.Ivanisenko,
S.S.Pintus,
D.A.Grigorovich,
and
N.A.Kolchanov
(2004).
PDBSiteScan: a program for searching for active, binding and posttranslational modification sites in the 3D structures of proteins.
|
| |
Nucleic Acids Res,
32,
W549-W554.
|
 |
|
|
|
|
 |
S.J.Lee,
S.J.Kim,
I.K.Kim,
J.Ko,
C.S.Jeong,
G.H.Kim,
C.Park,
S.O.Kang,
P.G.Suh,
H.S.Lee,
and
S.S.Cha
(2003).
Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain.
|
| |
J Biol Chem,
278,
44552-44559.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

