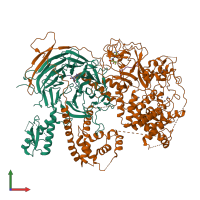Assemblies
Assembly Name:
Histone H3.1 and Polycomb Protein EED
Multimeric state:
hetero dimer
Accessible surface area:
22230.31 Å2
Buried surface area:
1043.16 Å2
Dissociation area:
521.58
Å2
Dissociation energy (ΔGdiss):
-0.36
kcal/mol
Dissociation entropy (TΔSdiss):
6.9
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-219065
Assembly Name:
SET domain-containing protein and Polycomb protein VEFS-Box domain-containing protein
Multimeric state:
hetero dimer
Accessible surface area:
46042.26 Å2
Buried surface area:
1575.25 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-219064
Macromolecules
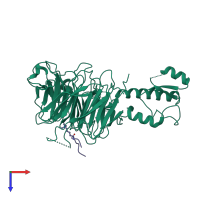
Chain: A
Length: 605 amino acids
Theoretical weight: 66.02 KDa
Source organism: Thermochaetoides thermophila DSM 1495
Expression system: Saccharomyces cerevisiae
UniProt:
Pfam: WD domain, G-beta repeat
InterPro:
Length: 605 amino acids
Theoretical weight: 66.02 KDa
Source organism: Thermochaetoides thermophila DSM 1495
Expression system: Saccharomyces cerevisiae
UniProt:
- Canonical:
 G0S8H7 (Residues: 1-565; Coverage: 100%)
G0S8H7 (Residues: 1-565; Coverage: 100%)
Pfam: WD domain, G-beta repeat
InterPro:
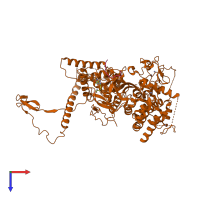
Chain: B
Length: 937 amino acids
Theoretical weight: 106.87 KDa
Source organism: Thermochaetoides thermophila DSM 1495
Expression system: Saccharomyces cerevisiae
UniProt:
Pfam:
Length: 937 amino acids
Theoretical weight: 106.87 KDa
Source organism: Thermochaetoides thermophila DSM 1495
Expression system: Saccharomyces cerevisiae
UniProt:
- Canonical:
 G0SDW4 (Residues: 191-952; Coverage: 50%)
G0SDW4 (Residues: 191-952; Coverage: 50%)
Pfam:
- EZH2 N-terminal domain
- Fungal MCSS domain
- Fungal polycomb repressive complex 2, Ezh2-like, preSET CXC domain
- SET domain
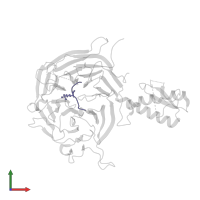
Chain: D
Length: 11 amino acids
Theoretical weight: 1.15 KDa
Source organism: Homo sapiens
Expression system: Not provided
UniProt:
Length: 11 amino acids
Theoretical weight: 1.15 KDa
Source organism: Homo sapiens
Expression system: Not provided
UniProt:
- Canonical:
 P68431 (Residues: 23-33; Coverage: 8%)
P68431 (Residues: 23-33; Coverage: 8%)
Chain: E
Length: 9 amino acids
Theoretical weight: 926 Da
Source organism: Thermochaetoides thermophila DSM 1495
Expression system: Saccharomyces cerevisiae
UniProt:
Length: 9 amino acids
Theoretical weight: 926 Da
Source organism: Thermochaetoides thermophila DSM 1495
Expression system: Saccharomyces cerevisiae
UniProt:
- Canonical:
 G0RYC6 (Residues: 531-539; Coverage: 1%)
G0RYC6 (Residues: 531-539; Coverage: 1%)