Assemblies
Assembly Name:
Transportin-1 and Histone H3.1
Multimeric state:
hetero dimer
Accessible surface area:
38446.56 Å2
Buried surface area:
1754.25 Å2
Dissociation area:
877.12
Å2
Dissociation energy (ΔGdiss):
0.16
kcal/mol
Dissociation entropy (TΔSdiss):
8.65
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-159434
Assembly Name:
Transportin-1 and Histone H3.1
Multimeric state:
hetero dimer
Accessible surface area:
38582.95 Å2
Buried surface area:
1844.48 Å2
Dissociation area:
922.24
Å2
Dissociation energy (ΔGdiss):
4.47
kcal/mol
Dissociation entropy (TΔSdiss):
8.61
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-159434
Macromolecules
Chains: A, B
Length: 854 amino acids
Theoretical weight: 96.77 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
CATH: Leucine-rich Repeat Variant
Length: 854 amino acids
Theoretical weight: 96.77 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q92973 (Residues: 9-331, 375-898; Coverage: 94%)
Q92973 (Residues: 9-331, 375-898; Coverage: 94%)
Pfam:
InterPro:
CATH: Leucine-rich Repeat Variant
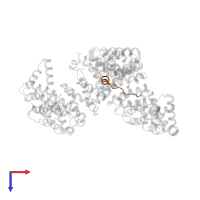
Chains: C, D
Length: 17 amino acids
Theoretical weight: 1.79 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
InterPro: Histone H3/CENP-A
Length: 17 amino acids
Theoretical weight: 1.79 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P68431 (Residues: 12-28; Coverage: 13%)
P68431 (Residues: 12-28; Coverage: 13%)
InterPro: Histone H3/CENP-A