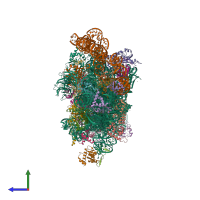
Assemblies
Multimeric state:
hetero 35-mer
Accessible surface area:
455926.43 Å2
Buried surface area:
202289.58 Å2
Dissociation area:
1,564.95
Å2
Dissociation energy (ΔGdiss):
2.62
kcal/mol
Dissociation entropy (TΔSdiss):
14.84
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-140329
Macromolecules
Chain: A
Length: 295 amino acids
Theoretical weight: 32.88 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam:
InterPro:
Length: 295 amino acids
Theoretical weight: 32.88 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P08865 (Residues: 1-295; Coverage: 100%)
P08865 (Residues: 1-295; Coverage: 100%)
Pfam:
InterPro:
- Small ribosomal subunit protein uS2, vertebrates
- Small ribosomal subunit protein uS2, eukaryotic
- Small ribosomal subunit protein uS2, eukaryotic/archaeal
- Small ribosomal subunit protein uS2, flavodoxin-like domain superfamily
- Small ribosomal subunit protein uS2
- Small ribosomal subunit protein uS2, conserved site
- Small ribosomal subunit protein uS2, C-terminal domain
Chain: B
Length: 264 amino acids
Theoretical weight: 30 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal S3Ae family
InterPro:
Length: 264 amino acids
Theoretical weight: 30 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P61247 (Residues: 1-264; Coverage: 100%)
P61247 (Residues: 1-264; Coverage: 100%)
Pfam: Ribosomal S3Ae family
InterPro:
Chain: C
Length: 293 amino acids
Theoretical weight: 31.38 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam:
InterPro:
Length: 293 amino acids
Theoretical weight: 31.38 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P15880 (Residues: 1-293; Coverage: 100%)
P15880 (Residues: 1-293; Coverage: 100%)
Pfam:
InterPro:
- Small ribosomal subunit protein uS5
- Small ribosomal subunit protein uS5, eukaryotic/archaeal
- Small ribosomal subunit protein uS5, N-terminal
- Small ribosomal subunit protein uS5, N-terminal, conserved site
- Small ribosomal subunit protein uS5 domain 2-type fold, subgroup
- Ribosomal protein uS5 domain 2-type superfamily
- Small ribosomal subunit protein uS5, C-terminal
Chain: D
Length: 243 amino acids
Theoretical weight: 26.73 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam:
InterPro:
Length: 243 amino acids
Theoretical weight: 26.73 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P23396 (Residues: 1-243; Coverage: 100%)
P23396 (Residues: 1-243; Coverage: 100%)
Pfam:
InterPro:
- K homology domain-like, alpha/beta
- Small ribosomal subunit protein uS3, eukaryotic/archaeal
- K homology domain superfamily, prokaryotic type
- K Homology domain, type 2
- Ribosomal protein S3, C-terminal domain superfamily
- Small ribosomal subunit protein uS3, C-terminal
- Small ribosomal subunit protein uS3, conserved site
Chain: E
Length: 263 amino acids
Theoretical weight: 29.51 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam:
InterPro:
Length: 263 amino acids
Theoretical weight: 29.51 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P22090 (Residues: 1-263; Coverage: 100%)
P22090 (Residues: 1-263; Coverage: 100%)
Pfam:
InterPro:
- Small ribosomal subunit protein eS4
- RNA-binding S4 domain superfamily
- Small ribosomal subunit protein eS4, N-terminal
- Small ribosomal subunit protein eS4, N-terminal, conserved site
- RNA-binding S4 domain
- Small ribosomal subunit protein eS4, central region
- Small ribosomal subunit protein eS4, central domain superfamily
- Large ribosomal subunit protein uL2, domain 2
- Small ribosomal subunit protein eS4, KOW domain
- KOW
- Small ribosomal subunit protein eS4, C-terminal domain
Chain: F
Length: 204 amino acids
Theoretical weight: 22.91 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S7p/S5e
InterPro:
Length: 204 amino acids
Theoretical weight: 22.91 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P46782 (Residues: 1-204; Coverage: 100%)
P46782 (Residues: 1-204; Coverage: 100%)
Pfam: Ribosomal protein S7p/S5e
InterPro:
- Small ribosomal subunit protein uS7
- Small ribosomal subunit protein uS7 domain superfamily
- Ribosomal protein uS7, eukaryotic/archaeal
- Small ribosomal subunit protein uS7 domain
- Small ribosomal subunit protein uS7, conserved site
Chain: G
Length: 249 amino acids
Theoretical weight: 28.75 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S6e
InterPro:
Length: 249 amino acids
Theoretical weight: 28.75 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62753 (Residues: 1-249; Coverage: 100%)
P62753 (Residues: 1-249; Coverage: 100%)
Pfam: Ribosomal protein S6e
InterPro:
- Small ribosomal subunit protein eS6
- Small ribosomal subunit protein eS6-like
- Small ribosomal subunit protein eS6, conserved site
Chain: H
Length: 194 amino acids
Theoretical weight: 22.17 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S7e
InterPro:
Length: 194 amino acids
Theoretical weight: 22.17 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62081 (Residues: 1-194; Coverage: 100%)
P62081 (Residues: 1-194; Coverage: 100%)
Pfam: Ribosomal protein S7e
InterPro:
Chain: I
Length: 208 amino acids
Theoretical weight: 24.26 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S8e
InterPro:
Length: 208 amino acids
Theoretical weight: 24.26 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62241 (Residues: 1-208; Coverage: 100%)
P62241 (Residues: 1-208; Coverage: 100%)
Pfam: Ribosomal protein S8e
InterPro:
- Small ribosomal subunit protein eS8
- Ribosomal protein eS8/ribosomal biogenesis NSA2
- Small ribosomal subunit protein eS8, conserved site
- Small ribosomal subunit protein eS8 subdomain, eukaryotes
Chain: J
Length: 194 amino acids
Theoretical weight: 22.64 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam:
InterPro:
Length: 194 amino acids
Theoretical weight: 22.64 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P46781 (Residues: 1-194; Coverage: 100%)
P46781 (Residues: 1-194; Coverage: 100%)
Pfam:
InterPro:
Chain: K
Length: 165 amino acids
Theoretical weight: 18.93 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Plectin/S10 domain
InterPro:
Length: 165 amino acids
Theoretical weight: 18.93 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P46783 (Residues: 1-165; Coverage: 100%)
P46783 (Residues: 1-165; Coverage: 100%)
Pfam: Plectin/S10 domain
InterPro:
- Winged helix-like DNA-binding domain superfamily
- Small ribosomal subunit protein eS10
- Plectin/eS10, N-terminal
Chain: L
Length: 158 amino acids
Theoretical weight: 18.47 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam:
InterPro:
Length: 158 amino acids
Theoretical weight: 18.47 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62280 (Residues: 1-158; Coverage: 100%)
P62280 (Residues: 1-158; Coverage: 100%)
Pfam:
InterPro:
- Small ribosomal subunit protein uS17, N-terminal
- Small ribosomal subunit protein uS17
- Small ribosomal subunit protein uS17, archaeal/eukaryotic
- Nucleic acid-binding, OB-fold
- Small ribosomal subunit protein uS17, conserved site
Chain: M
Length: 132 amino acids
Theoretical weight: 14.54 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein L7Ae/L30e/S12e/Gadd45 family
InterPro:
Length: 132 amino acids
Theoretical weight: 14.54 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P25398 (Residues: 1-132; Coverage: 100%)
P25398 (Residues: 1-132; Coverage: 100%)
Pfam: Ribosomal protein L7Ae/L30e/S12e/Gadd45 family
InterPro:
- Ribosomal protein eL30-like superfamily
- Ribosomal protein eL8/eL30/eS12/Gadd45
- Small ribosomal subunit protein eS12, conserved site
- Small ribosomal subunit protein eS12
Chain: N
Length: 151 amino acids
Theoretical weight: 17.26 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam:
InterPro:
Length: 151 amino acids
Theoretical weight: 17.26 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62277 (Residues: 1-151; Coverage: 100%)
P62277 (Residues: 1-151; Coverage: 100%)
Pfam:
InterPro:
- Small ribosomal subunit protein uS15, N-terminal
- Small ribosomal subunit protein uS15, archaea/eukaryotes
- uS15/NS1, RNA-binding domain superfamily
- Small ribosomal subunit protein uS15
Chain: O
Length: 151 amino acids
Theoretical weight: 16.3 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S11
InterPro:
Length: 151 amino acids
Theoretical weight: 16.3 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62263 (Residues: 1-151; Coverage: 100%)
P62263 (Residues: 1-151; Coverage: 100%)
Pfam: Ribosomal protein S11
InterPro:
- Small ribosomal subunit protein uS11
- Small ribosomal subunit protein uS11 superfamily
- Small ribosomal subunit protein uS11, conserved site
Chain: P
Length: 145 amino acids
Theoretical weight: 17.08 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S19
InterPro:
Length: 145 amino acids
Theoretical weight: 17.08 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62841 (Residues: 1-145; Coverage: 100%)
P62841 (Residues: 1-145; Coverage: 100%)
Pfam: Ribosomal protein S19
InterPro:
Chain: Q
Length: 146 amino acids
Theoretical weight: 16.48 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S9/S16
InterPro:
Length: 146 amino acids
Theoretical weight: 16.48 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62249 (Residues: 1-146; Coverage: 100%)
P62249 (Residues: 1-146; Coverage: 100%)
Pfam: Ribosomal protein S9/S16
InterPro:
- Small ribosomal subunit protein uS5 domain 2-type fold, subgroup
- Ribosomal protein uS5 domain 2-type superfamily
- Small ribosomal subunit protein uS9
- Small ribosomal subunit protein uS9, conserved site
Chain: R
Length: 135 amino acids
Theoretical weight: 15.58 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal S17
InterPro:
Length: 135 amino acids
Theoretical weight: 15.58 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P08708 (Residues: 1-135; Coverage: 100%)
P08708 (Residues: 1-135; Coverage: 100%)
Pfam: Ribosomal S17
InterPro:
- Small ribosomal subunit protein eS17
- Small ribosomal subunit protein eS17 superfamily
- Small ribosomal subunit protein eS17, conserved site
Chain: S
Length: 152 amino acids
Theoretical weight: 17.76 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S13/S18
InterPro:
Length: 152 amino acids
Theoretical weight: 17.76 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62269 (Residues: 1-152; Coverage: 100%)
P62269 (Residues: 1-152; Coverage: 100%)
Pfam: Ribosomal protein S13/S18
InterPro:
- Small ribosomal subunit protein uS13
- Small ribosomal subunit protein uS13-like, H2TH
- Small ribosomal subunit protein uS13, C-terminal
- Small ribosomal subunit protein uS13, conserved site
Chain: T
Length: 145 amino acids
Theoretical weight: 16.09 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S19e
InterPro:
Length: 145 amino acids
Theoretical weight: 16.09 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P39019 (Residues: 1-145; Coverage: 100%)
P39019 (Residues: 1-145; Coverage: 100%)
Pfam: Ribosomal protein S19e
InterPro:
Chain: U
Length: 119 amino acids
Theoretical weight: 13.4 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S10p/S20e
InterPro:
Length: 119 amino acids
Theoretical weight: 13.4 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P60866 (Residues: 1-119; Coverage: 100%)
P60866 (Residues: 1-119; Coverage: 100%)
Pfam: Ribosomal protein S10p/S20e
InterPro:
- Small ribosomal subunit protein uS10 domain superfamily
- Small ribosomal subunit protein uS10
- Small ribosomal subunit protein uS10, eukaryotic/archaeal
- Small ribosomal subunit protein uS10 domain
- Small ribosomal subunit protein uS10, conserved site
Chain: V
Length: 83 amino acids
Theoretical weight: 9.12 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S21e
InterPro:
Length: 83 amino acids
Theoretical weight: 9.12 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P63220 (Residues: 1-83; Coverage: 100%)
P63220 (Residues: 1-83; Coverage: 100%)
Pfam: Ribosomal protein S21e
InterPro:
- Small ribosomal subunit protein eS21
- Small ribosomal subunit protein eS21 superfamily
- Small ribosomal subunit protein eS21, conserved site
Chain: W
Length: 130 amino acids
Theoretical weight: 14.87 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S8
InterPro:
Length: 130 amino acids
Theoretical weight: 14.87 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62244 (Residues: 1-130; Coverage: 100%)
P62244 (Residues: 1-130; Coverage: 100%)
Pfam: Ribosomal protein S8
InterPro:
- Small ribosomal subunit protein uS8
- Small ribosomal subunit protein uS8 superfamily
- Small ribosomal subunit protein uS8, conserved site
Chain: X
Length: 143 amino acids
Theoretical weight: 15.84 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S12/S23
InterPro:
Length: 143 amino acids
Theoretical weight: 15.84 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62266 (Residues: 1-143; Coverage: 100%)
P62266 (Residues: 1-143; Coverage: 100%)
Pfam: Ribosomal protein S12/S23
InterPro:
Chain: Y
Length: 133 amino acids
Theoretical weight: 15.46 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S24e
InterPro:
Length: 133 amino acids
Theoretical weight: 15.46 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62847 (Residues: 1-133; Coverage: 100%)
P62847 (Residues: 1-133; Coverage: 100%)
Pfam: Ribosomal protein S24e
InterPro:
- Small ribosomal subunit protein eS24
- Ribosomal protein uL23/eL15/eS24 core domain superfamily
- Small ribosomal subunit protein eS24 conserved site
Chain: Z
Length: 125 amino acids
Theoretical weight: 13.78 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: S25 ribosomal protein
InterPro:
CATH: Winged helix-like DNA-binding domain superfamily/Winged helix DNA-binding domain
Length: 125 amino acids
Theoretical weight: 13.78 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62851 (Residues: 1-125; Coverage: 100%)
P62851 (Residues: 1-125; Coverage: 100%)
Pfam: S25 ribosomal protein
InterPro:
CATH: Winged helix-like DNA-binding domain superfamily/Winged helix DNA-binding domain
Chain: a
Length: 115 amino acids
Theoretical weight: 13.05 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S26e
InterPro:
Length: 115 amino acids
Theoretical weight: 13.05 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62854 (Residues: 1-115; Coverage: 100%)
P62854 (Residues: 1-115; Coverage: 100%)
Pfam: Ribosomal protein S26e
InterPro:
- Small ribosomal subunit protein eS26
- Ribosomal protein eS26 superfamily
- Ribosomal protein eS26, conserved site
Chain: b
Length: 84 amino acids
Theoretical weight: 9.48 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S27
InterPro:
Length: 84 amino acids
Theoretical weight: 9.48 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P42677 (Residues: 1-84; Coverage: 100%)
P42677 (Residues: 1-84; Coverage: 100%)
Pfam: Ribosomal protein S27
InterPro:
- Small ribosomal subunit protein eS27
- Small ribosomal subunit protein eS27, zinc-binding domain superfamily
- Zinc-binding ribosomal protein
Chain: c
Length: 69 amino acids
Theoretical weight: 7.86 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S28e
InterPro:
Length: 69 amino acids
Theoretical weight: 7.86 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62857 (Residues: 1-69; Coverage: 100%)
P62857 (Residues: 1-69; Coverage: 100%)
Pfam: Ribosomal protein S28e
InterPro:
- Nucleic acid-binding, OB-fold
- Small ribosomal subunit protein eS28
- Ribosomal protein eS28 conserved site
Chain: d
Length: 56 amino acids
Theoretical weight: 6.69 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S14p/S29e
InterPro:
Length: 56 amino acids
Theoretical weight: 6.69 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62273 (Residues: 1-56; Coverage: 100%)
P62273 (Residues: 1-56; Coverage: 100%)
Pfam: Ribosomal protein S14p/S29e
InterPro:
- Small ribosomal subunit protein uS14, eukaryotes/archaea
- Small ribosomal subunit protein uS14 superfamily
- Small ribosomal subunit protein uS14
- Small ribosomal subunit protein uS14, conserved site
Chain: e
Length: 59 amino acids
Theoretical weight: 6.67 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: Ribosomal protein S30
InterPro: Small ribosomal subunit protein eS30
Length: 59 amino acids
Theoretical weight: 6.67 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62861 (Residues: 75-133; Coverage: 44%)
P62861 (Residues: 75-133; Coverage: 44%)
Pfam: Ribosomal protein S30
InterPro: Small ribosomal subunit protein eS30
Chain: f
Length: 156 amino acids
Theoretical weight: 18 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam:
InterPro:
Length: 156 amino acids
Theoretical weight: 18 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P62979 (Residues: 1-156; Coverage: 100%)
P62979 (Residues: 1-156; Coverage: 100%)
Pfam:
InterPro:
- Zinc-binding ribosomal protein
- Ubiquitin-like domain
- Ubiquitin-like domain superfamily
- Ubiquitin domain
- Ubiquitin conserved site
- Small ribosomal subunit protein eS31 eukaryotic-type superfamily
- Small ribosomal subunit protein eS31
Chain: g
Length: 317 amino acids
Theoretical weight: 35.12 KDa
Source organism: Oryctolagus cuniculus
UniProt:
Pfam: WD domain, G-beta repeat
InterPro:
Length: 317 amino acids
Theoretical weight: 35.12 KDa
Source organism: Oryctolagus cuniculus
UniProt:
- Canonical:
 P63244 (Residues: 1-317; Coverage: 100%)
P63244 (Residues: 1-317; Coverage: 100%)
Pfam: WD domain, G-beta repeat
InterPro:
- Small ribosomal subunit protein RACK1-like
- WD40 repeat
- WD40-repeat-containing domain superfamily
- WD40/YVTN repeat-like-containing domain superfamily
- WD40 repeat, conserved site
- G-protein beta WD-40 repeat
Name:
18S Ribosomal RNA
Representative chains: 1
Length: 1869 nucleotides
Theoretical weight: 602.78 KDa
Rfam: Eukaryotic small subunit ribosomal RNA
Representative chains: 1
Length: 1869 nucleotides
Theoretical weight: 602.78 KDa
Rfam: Eukaryotic small subunit ribosomal RNA
Name:
HCV-IRES
Representative chains: z
Length: 504 nucleotides
Theoretical weight: 162.19 KDa
Rfam:
Hepatitis C virus internal ribosome entry site
Hepatitis C alternative reading frame stem-loop
Representative chains: z
Length: 504 nucleotides
Theoretical weight: 162.19 KDa
Rfam: