Assemblies
Assembly Name:
DNA topoisomerase 4 subunit B
Multimeric state:
monomeric
Accessible surface area:
9437.94 Å2
Buried surface area:
0.0 Å2
Dissociation area:
0
Å2
Dissociation energy (ΔGdiss):
0
kcal/mol
Dissociation entropy (TΔSdiss):
0
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-184153
Macromolecules
Chain: A
Length: 226 amino acids
Theoretical weight: 24.6 KDa
Source organism: Streptococcus pneumoniae R6
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase
InterPro:
Length: 226 amino acids
Theoretical weight: 24.6 KDa
Source organism: Streptococcus pneumoniae R6
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q8DQB5 (Residues: 1-226; Coverage: 35%)
Q8DQB5 (Residues: 1-226; Coverage: 35%)
Pfam: Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase
InterPro:
- Histidine kinase/HSP90-like ATPase superfamily
- DNA topoisomerase, type IIA, subunit B
- Histidine kinase/HSP90-like ATPase
- DNA topoisomerase, type IIA



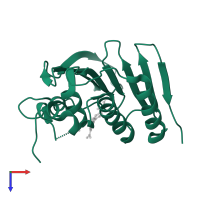
![The deposited structure of PDB entry <span class='highlight'>4lp0</span> coloured by chain, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>DNA topoisomerase 4 subunit B</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 4lp0 coloured by chain, front view.](https://www.ebi.ac.uk/pdbe/static/entry/4lp0_deposited_chain_front_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>4lp0</span> coloured by chain, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>DNA topoisomerase 4 subunit B</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 4lp0 coloured by chain, side view.](https://www.ebi.ac.uk/pdbe/static/entry/4lp0_deposited_chain_side_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>4lp0</span> coloured by chain, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>DNA topoisomerase 4 subunit B</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul> PDB entry 4lp0 coloured by chain, top view.](https://www.ebi.ac.uk/pdbe/static/entry/4lp0_deposited_chain_top_image-200x200.png)
![Monomeric assembly 1 of PDB entry <span class='highlight'>4lp0</span> coloured by chemically distinct molecules, front view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>DNA topoisomerase 4 subunit B</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 1 of PDB entry 4lp0 coloured by chemically distinct molecules, front view.](https://www.ebi.ac.uk/pdbe/static/entry/4lp0_assembly_1_chemically_distinct_molecules_front_image-200x200.png)
![Monomeric assembly 1 of PDB entry <span class='highlight'>4lp0</span> coloured by chemically distinct molecules, side view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>DNA topoisomerase 4 subunit B</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 1 of PDB entry 4lp0 coloured by chemically distinct molecules, side view.](https://www.ebi.ac.uk/pdbe/static/entry/4lp0_assembly_1_chemically_distinct_molecules_side_image-200x200.png)
![Monomeric assembly 1 of PDB entry <span class='highlight'>4lp0</span> coloured by chemically distinct molecules, top view. This structure contains: <ul class='image_legend_ul'><li class='image_legend_li'>1 copy of <span class='highlight'>DNA topoisomerase 4 subunit B</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid</span>;</li> <li class='image_legend_li'>1 copy of <span class='highlight'>water</span>.</li></ul>} Monomeric assembly 1 of PDB entry 4lp0 coloured by chemically distinct molecules, top view.](https://www.ebi.ac.uk/pdbe/static/entry/4lp0_assembly_1_chemically_distinct_molecules_top_image-200x200.png)