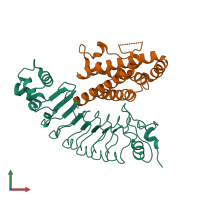
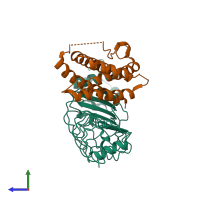
Assemblies
Multimeric state:
hetero dimer
Accessible surface area:
19737.97 Å2
Buried surface area:
1982.09 Å2
Dissociation area:
991.04
Å2
Dissociation energy (ΔGdiss):
-2.17
kcal/mol
Dissociation entropy (TΔSdiss):
12.36
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-107391
Multimeric state:
hetero dimer
Accessible surface area:
19590.47 Å2
Buried surface area:
1896.37 Å2
Dissociation area:
948.19
Å2
Dissociation energy (ΔGdiss):
-2.78
kcal/mol
Dissociation entropy (TΔSdiss):
12.4
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-107391
Macromolecules
Chains: A, B
Length: 267 amino acids
Theoretical weight: 30.06 KDa
Source organisms: Expression system: Escherichia coli
UniProt:
Pfam:
InterPro:
Length: 267 amino acids
Theoretical weight: 30.06 KDa
Source organisms: Expression system: Escherichia coli
UniProt:
- Canonical:
 A4L9V2 (Residues: 43-122, 145-153; Coverage: 14%)
A4L9V2 (Residues: 43-122, 145-153; Coverage: 14%) - Canonical:
 Q4G1L3 (Residues: 54-77, 87-232; Coverage: 60%)
nullnull
Q4G1L3 (Residues: 54-77, 87-232; Coverage: 60%)
nullnull
Pfam:
InterPro:
- Leucine-rich repeat domain superfamily
- Leucine-rich repeat, typical subtype
- Leucine-rich repeat
- Variable lymphocyte receptor, C-terminal
Chains: C, D
Length: 168 amino acids
Theoretical weight: 19.2 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Interleukin-6/G-CSF/MGF family
InterPro:
Length: 168 amino acids
Theoretical weight: 19.2 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P05231 (Residues: 47-212; Coverage: 91%)
P05231 (Residues: 47-212; Coverage: 91%)
Pfam: Interleukin-6/G-CSF/MGF family
InterPro:
- Four-helical cytokine-like, core
- Interleukin-6-like
- Interleukin-6/GCSF/MGF
- Interleukin-6/GCSF/MGF, conserved site