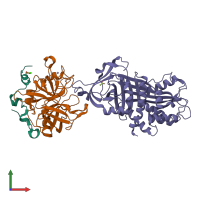
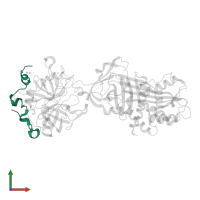
Assemblies
Assembly Name:
alpha-thrombin complex and Glia-derived nexin
Multimeric state:
hetero trimer
Accessible surface area:
26311.97 Å2
Buried surface area:
5859.15 Å2
Dissociation area:
783.1
Å2
Dissociation energy (ΔGdiss):
3.45
kcal/mol
Dissociation entropy (TΔSdiss):
12.97
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-133071
Assembly Name:
alpha-thrombin complex and Glia-derived nexin
Multimeric state:
hetero trimer
Accessible surface area:
27545.13 Å2
Buried surface area:
3900.22 Å2
Dissociation area:
1,037.53
Å2
Dissociation energy (ΔGdiss):
0.88
kcal/mol
Dissociation entropy (TΔSdiss):
12.94
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-133071
Macromolecules
Chains: A, D
Length: 49 amino acids
Theoretical weight: 5.64 KDa
Source organism: Homo sapiens
Expression system: Cricetinae gen. sp.
UniProt:
Pfam: Thrombin light chain
InterPro:
CATH: Thrombin light chain domain
Length: 49 amino acids
Theoretical weight: 5.64 KDa
Source organism: Homo sapiens
Expression system: Cricetinae gen. sp.
UniProt:
- Canonical:
 P00734 (Residues: 315-363; Coverage: 8%)
P00734 (Residues: 315-363; Coverage: 8%)
Pfam: Thrombin light chain
InterPro:
CATH: Thrombin light chain domain
Chains: B, E
Length: 259 amino acids
Theoretical weight: 29.76 KDa
Source organism: Homo sapiens
Expression system: Cricetinae gen. sp.
UniProt:
Pfam: Trypsin
InterPro:
Length: 259 amino acids
Theoretical weight: 29.76 KDa
Source organism: Homo sapiens
Expression system: Cricetinae gen. sp.
UniProt:
- Canonical:
 P00734 (Residues: 364-622; Coverage: 43%)
P00734 (Residues: 364-622; Coverage: 43%)
Pfam: Trypsin
InterPro:
- Serine proteases, trypsin domain
- Peptidase S1, PA clan
- Peptidase S1, PA clan, chymotrypsin-like fold
- Peptidase S1A, chymotrypsin family
- Serine proteases, trypsin family, histidine active site
- Prothrombin/thrombin
Chains: C, F
Length: 379 amino acids
Theoretical weight: 41.92 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Serpin (serine protease inhibitor)
InterPro:
Length: 379 amino acids
Theoretical weight: 41.92 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P07093 (Residues: 20-398; Coverage: 100%)
P07093 (Residues: 20-398; Coverage: 100%)
Pfam: Serpin (serine protease inhibitor)
InterPro:
- Serpin superfamily
- Serpin domain
- Serpin family
- Serpin superfamily, domain 1
- Serpin superfamily, domain 2
- Serpin, conserved site