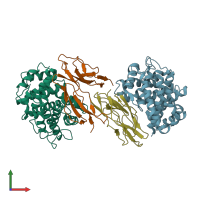
Assemblies
Assembly Name:
Complement C3b complex and Complement receptor type 2
Multimeric state:
hetero dimer
Accessible surface area:
18259.22 Å2
Buried surface area:
2241.21 Å2
Dissociation area:
1,120.61
Å2
Dissociation energy (ΔGdiss):
-6.45
kcal/mol
Dissociation entropy (TΔSdiss):
12.3
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-133844
Assembly Name:
Complement C3b complex and Complement receptor type 2
Multimeric state:
hetero dimer
Accessible surface area:
18569.21 Å2
Buried surface area:
2150.16 Å2
Dissociation area:
1,075.08
Å2
Dissociation energy (ΔGdiss):
-6.22
kcal/mol
Dissociation entropy (TΔSdiss):
12.3
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-133844
Macromolecules
Chains: A, B
Length: 310 amino acids
Theoretical weight: 34.75 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21
UniProt:
Pfam: A-macroglobulin TED domain
InterPro:
Length: 310 amino acids
Theoretical weight: 34.75 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21
UniProt:
- Canonical:
 P01024 (Residues: 996-1303; Coverage: 19%)
P01024 (Residues: 996-1303; Coverage: 19%)
Pfam: A-macroglobulin TED domain
InterPro:
- Terpenoid cyclases/protein prenyltransferase alpha-alpha toroid
- Alpha-macroglobulin-like, TED domain
- Alpha-macroglobulin-like, thiol-ester bond-forming region
Chains: C, D
Length: 135 amino acids
Theoretical weight: 14.83 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21
UniProt:
Pfam: Sushi repeat (SCR repeat)
InterPro:
CATH: Complement Module, domain 1
Length: 135 amino acids
Theoretical weight: 14.83 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21
UniProt:
- Canonical:
 P20023 (Residues: 19-153; Coverage: 13%)
P20023 (Residues: 19-153; Coverage: 13%)
Pfam: Sushi repeat (SCR repeat)
InterPro:
CATH: Complement Module, domain 1