EC 1.2.4.1: Pyruvate dehydrogenase (acetyl-transferring)
Reaction catalysed:
Pyruvate + [dihydrolipoyllysine-residue acetyltransferase] lipoyllysine = [dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine + CO(2)
Systematic name:
Pyruvate:[dihydrolipoyllysine-residue acetyltransferase]-lipoyllysine 2-oxidoreductase (decarboxylating, acceptor-acetylating)
Alternative Name(s):
- PDH
- Pyruvate decarboxylase
- Pyruvate dehydrogenase
- Pyruvate dehydrogenase (lipoamide)
- Pyruvate dehydrogenase complex
- Pyruvate:lipoamide 2-oxidoreductase (decarboxylating and acceptor-acetylating)
- Pyruvic acid dehydrogenase
- Pyruvic dehydrogenase
GO terms
Biochemical function:
Biological process:
Cellular component:
Sequence families
 Protein families (Pfam)
Protein families (Pfam)
 InterPro annotations
InterPro annotations
IPR009014
Domain description:
Transketolase C-terminal/Pyruvate-ferredoxin oxidoreductase domain II
Occurring in:
Occurring in:



![The deposited structure of PDB entry <span class='highlight'>3exg</span> contains 16 copies of Pfam domain <span class='highlight'>PF02780 (Transketolase, C-terminal domain)</span> in <span class='highlight'>Pyruvate dehydrogenase E1 component subunit beta, mitochondrial</span>. Showing 1 copy in chain <span class='highlight'>BA [auth 2]</span>. The deposited structure of PDB entry 3exg contains 16 copies of Pfam domain PF02780 (Transketolase, C-terminal domain) in Pyruvate dehydrogenase E1 component subunit beta, mitochondrial. Showing 1 copy in chain BA [auth 2].](https://www.ebi.ac.uk/pdbe/static/entry/3exg_2_2_Pfam_PF02780_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>3exg</span> contains 16 copies of Pfam domain <span class='highlight'>PF02779 (Transketolase, pyrimidine binding domain)</span> in <span class='highlight'>Pyruvate dehydrogenase E1 component subunit beta, mitochondrial</span>. Showing 1 copy in chain <span class='highlight'>BA [auth 2]</span>. The deposited structure of PDB entry 3exg contains 16 copies of Pfam domain PF02779 (Transketolase, pyrimidine binding domain) in Pyruvate dehydrogenase E1 component subunit beta, mitochondrial. Showing 1 copy in chain BA [auth 2].](https://www.ebi.ac.uk/pdbe/static/entry/3exg_2_2_Pfam_PF02779_image-200x200.png)

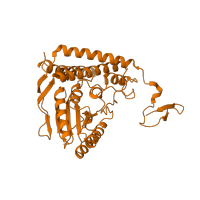
![The deposited structure of PDB entry <span class='highlight'>3exg</span> contains 16 copies of CATH domain <span class='highlight'>3.40.50.970 (Rossmann fold)</span> in <span class='highlight'>Pyruvate dehydrogenase E1 component subunit beta, mitochondrial</span>. Showing 1 copy in chain <span class='highlight'>BA [auth 2]</span>. The deposited structure of PDB entry 3exg contains 16 copies of CATH domain 3.40.50.970 (Rossmann fold) in Pyruvate dehydrogenase E1 component subunit beta, mitochondrial. Showing 1 copy in chain BA [auth 2].](https://www.ebi.ac.uk/pdbe/static/entry/3exg_2_2_CATH_3.40.50.970_image-200x200.png)
![The deposited structure of PDB entry <span class='highlight'>3exg</span> contains 16 copies of CATH domain <span class='highlight'>3.40.50.920 (Rossmann fold)</span> in <span class='highlight'>Pyruvate dehydrogenase E1 component subunit beta, mitochondrial</span>. Showing 1 copy in chain <span class='highlight'>BA [auth 2]</span>. The deposited structure of PDB entry 3exg contains 16 copies of CATH domain 3.40.50.920 (Rossmann fold) in Pyruvate dehydrogenase E1 component subunit beta, mitochondrial. Showing 1 copy in chain BA [auth 2].](https://www.ebi.ac.uk/pdbe/static/entry/3exg_2_2_CATH_3.40.50.920_image-200x200.png)