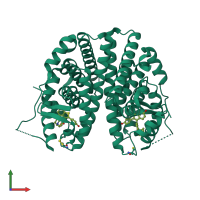
Assemblies
Assembly Name:
Estrogen receptor
Multimeric state:
homo dimer
Accessible surface area:
19953.68 Å2
Buried surface area:
5122.69 Å2
Dissociation area:
1,490.97
Å2
Dissociation energy (ΔGdiss):
3.14
kcal/mol
Dissociation entropy (TΔSdiss):
13.23
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-136919
Macromolecules
Chains: A, B
Length: 258 amino acids
Theoretical weight: 29.51 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Ligand-binding domain of nuclear hormone receptor
InterPro:
Length: 258 amino acids
Theoretical weight: 29.51 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 P03372 (Residues: 298-554; Coverage: 43%)
P03372 (Residues: 298-554; Coverage: 43%)
Pfam: Ligand-binding domain of nuclear hormone receptor
InterPro:
- Nuclear hormone receptor-like domain superfamily
- Nuclear hormone receptor, ligand-binding domain
- Nuclear hormone receptor