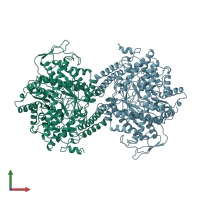
Assemblies
Assembly Name:
Formate acetyltransferase 1
Multimeric state:
homo dimer
Accessible surface area:
48858.8 Å2
Buried surface area:
4367.84 Å2
Dissociation area:
1,904.22
Å2
Dissociation energy (ΔGdiss):
11.64
kcal/mol
Dissociation entropy (TΔSdiss):
15.6
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-140621
Macromolecules
Chains: A, B
Length: 759 amino acids
Theoretical weight: 85.33 KDa
Source organism: Escherichia coli
UniProt:
Pfam:
InterPro:
SCOP: PFL-like
Length: 759 amino acids
Theoretical weight: 85.33 KDa
Source organism: Escherichia coli
UniProt:
- Canonical:
 P09373 (Residues: 2-760; Coverage: 100%)
P09373 (Residues: 2-760; Coverage: 100%)
Pfam:
InterPro:
- Pyruvate formate lyase domain
- Formate acetyltransferase
- Glycine radical domain
- Formate C-acetyltransferase glycine radical, conserved site
SCOP: PFL-like