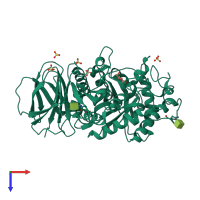
Assemblies
Assembly Name:
Lysosomal acid glucosylceramidase
Multimeric state:
monomeric
Accessible surface area:
18692.42 Å2
Buried surface area:
1298.27 Å2
Dissociation area:
136.98
Å2
Dissociation energy (ΔGdiss):
0.97
kcal/mol
Dissociation entropy (TΔSdiss):
0.84
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-137377
Assembly Name:
Lysosomal acid glucosylceramidase
Multimeric state:
monomeric
Accessible surface area:
19016.25 Å2
Buried surface area:
1811.95 Å2
Dissociation area:
90.35
Å2
Dissociation energy (ΔGdiss):
-5.62
kcal/mol
Dissociation entropy (TΔSdiss):
2.96
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-137377
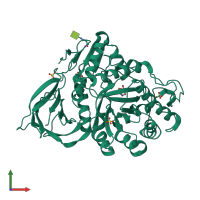
Assembly Name:
Lysosomal acid glucosylceramidase
Multimeric state:
monomeric
Accessible surface area:
18439.78 Å2
Buried surface area:
1084.6 Å2
Dissociation area:
141.5
Å2
Dissociation energy (ΔGdiss):
0.65
kcal/mol
Dissociation entropy (TΔSdiss):
0.89
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-137377
Assembly Name:
Lysosomal acid glucosylceramidase
Multimeric state:
monomeric
Accessible surface area:
18744.88 Å2
Buried surface area:
1311.61 Å2
Dissociation area:
114.34
Å2
Dissociation energy (ΔGdiss):
-5.56
kcal/mol
Dissociation entropy (TΔSdiss):
2.94
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-137377
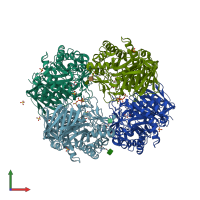
Assembly Name:
Lysosomal acid glucosylceramidase
Multimeric state:
homo tetramer
Accessible surface area:
69850.66 Å2
Buried surface area:
10549.1 Å2
Dissociation area:
982.45
Å2
Dissociation energy (ΔGdiss):
7.82
kcal/mol
Dissociation entropy (TΔSdiss):
15.52
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-137379
Assembly Name:
Lysosomal acid glucosylceramidase
Multimeric state:
homo tetramer
Accessible surface area:
67709.13 Å2
Buried surface area:
12690.64 Å2
Dissociation area:
2,090.3
Å2
Dissociation energy (ΔGdiss):
1.01
kcal/mol
Dissociation entropy (TΔSdiss):
15.81
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-137379
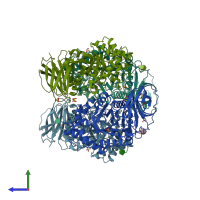
Assembly Name:
Lysosomal acid glucosylceramidase
Multimeric state:
homo dimer
Accessible surface area:
36137.36 Å2
Buried surface area:
4681.53 Å2
Dissociation area:
785.65
Å2
Dissociation energy (ΔGdiss):
4.06
kcal/mol
Dissociation entropy (TΔSdiss):
14.58
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-137378
Assembly Name:
Lysosomal acid glucosylceramidase
Multimeric state:
homo dimer
Accessible surface area:
35693.94 Å2
Buried surface area:
3886.93 Å2
Dissociation area:
745.36
Å2
Dissociation energy (ΔGdiss):
3.46
kcal/mol
Dissociation entropy (TΔSdiss):
14.56
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-137378
Assembly Name:
Lysosomal acid glucosylceramidase
Multimeric state:
homo dimer
Accessible surface area:
35915.32 Å2
Buried surface area:
3599.75 Å2
Dissociation area:
600.56
Å2
Dissociation energy (ΔGdiss):
0.64
kcal/mol
Dissociation entropy (TΔSdiss):
14.32
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-137378
Macromolecules
Chains: A, B, C, D
Length: 497 amino acids
Theoretical weight: 55.64 KDa
Source organism: Homo sapiens
Expression system: Cricetulus griseus
UniProt:
Pfam:
InterPro:
SCOP:
Length: 497 amino acids
Theoretical weight: 55.64 KDa
Source organism: Homo sapiens
Expression system: Cricetulus griseus
UniProt:
- Canonical:
 P04062 (Residues: 40-536; Coverage: 100%)
P04062 (Residues: 40-536; Coverage: 100%)
Pfam:
InterPro:
- Glycoside hydrolase family 30
- Glycosyl hydrolase family 30, TIM-barrel domain
- Glycoside hydrolase superfamily
- Glycosyl hydrolase family 30, beta sandwich domain
SCOP: