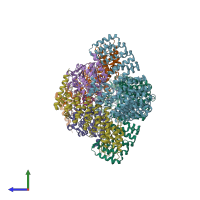
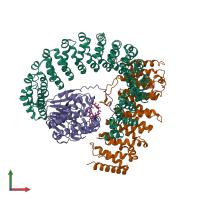
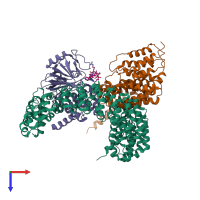
Assemblies
Multimeric state:
hetero tetramer
Accessible surface area:
54649.83 Å2
Buried surface area:
8217.61 Å2
Dissociation area:
3,985.77
Å2
Dissociation energy (ΔGdiss):
8.26
kcal/mol
Dissociation entropy (TΔSdiss):
36.37
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-151657
Multimeric state:
hetero tetramer
Accessible surface area:
54742.57 Å2
Buried surface area:
8195.65 Å2
Dissociation area:
3,974.06
Å2
Dissociation energy (ΔGdiss):
6.59
kcal/mol
Dissociation entropy (TΔSdiss):
36.37
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-151657
Macromolecules
Chains: A, D
Length: 589 amino acids
Theoretical weight: 65.38 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: HEAT repeat
InterPro:
CATH: Leucine-rich Repeat Variant
SCOP: HEAT repeat
Length: 589 amino acids
Theoretical weight: 65.38 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 P30153 (Residues: 1-589; Coverage: 100%)
P30153 (Residues: 1-589; Coverage: 100%)
Pfam: HEAT repeat
InterPro:
CATH: Leucine-rich Repeat Variant
SCOP: HEAT repeat
Chains: B, E
Length: 449 amino acids
Theoretical weight: 52.7 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Protein phosphatase 2A regulatory B subunit (B56 family)
InterPro:
CATH: Leucine-rich Repeat Variant
SCOP: B56-like
Length: 449 amino acids
Theoretical weight: 52.7 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q13362 (Residues: 1-442; Coverage: 84%)
Q13362 (Residues: 1-442; Coverage: 84%)
Pfam: Protein phosphatase 2A regulatory B subunit (B56 family)
InterPro:
CATH: Leucine-rich Repeat Variant
SCOP: B56-like
Chains: C, F
Length: 309 amino acids
Theoretical weight: 35.64 KDa
Source organism: Homo sapiens
Expression system: Spodoptera frugiperda
UniProt:
Pfam: Calcineurin-like phosphoesterase
InterPro:
SCOP: Protein serine/threonine phosphatase
Length: 309 amino acids
Theoretical weight: 35.64 KDa
Source organism: Homo sapiens
Expression system: Spodoptera frugiperda
UniProt:
- Canonical:
 P67775 (Residues: 1-309; Coverage: 100%)
P67775 (Residues: 1-309; Coverage: 100%)
Pfam: Calcineurin-like phosphoesterase
InterPro:
- Metallo-dependent phosphatase-like
- Serine/Threonine protein phosphatase PP2A-like
- Serine/threonine-specific protein phosphatase/bis(5-nucleosyl)-tetraphosphatase
- Calcineurin-like phosphoesterase domain, ApaH type
SCOP: Protein serine/threonine phosphatase
Chains: X, Y
Length: 7 amino acids
Theoretical weight: 1.01 KDa
Source organism: Cyanobacteriota
Expression system: Not provided
Length: 7 amino acids
Theoretical weight: 1.01 KDa
Source organism: Cyanobacteriota
Expression system: Not provided