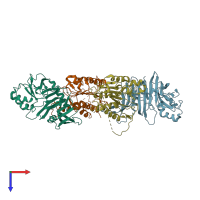
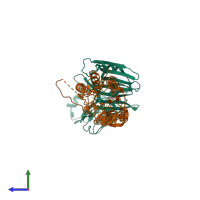
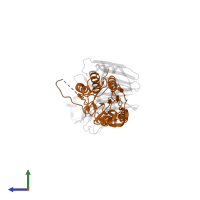
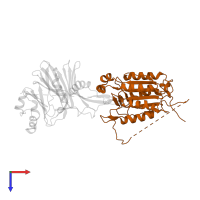
Assemblies
Assembly Name:
Early 35 kDa protein and Caspase-8 subunit p18
Multimeric state:
hetero dimer
Accessible surface area:
26452.18 Å2
Buried surface area:
1873.17 Å2
Dissociation area:
936.58
Å2
Dissociation energy (ΔGdiss):
-0.71
kcal/mol
Dissociation entropy (TΔSdiss):
12.84
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-139942
Assembly Name:
Early 35 kDa protein and Caspase-8 subunit p18
Multimeric state:
hetero dimer
Accessible surface area:
25514.28 Å2
Buried surface area:
1951.83 Å2
Dissociation area:
975.91
Å2
Dissociation energy (ΔGdiss):
-1.72
kcal/mol
Dissociation entropy (TΔSdiss):
12.84
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-139942
Macromolecules
Chains: A, C
Length: 298 amino acids
Theoretical weight: 34.74 KDa
Source organism: Autographa californica nucleopolyhedrovirus
Expression system: Escherichia coli
UniProt:
Pfam: Apoptosis preventing protein
InterPro:
CATH: Baculovirus p35
SCOP: Baculovirus p35 protein
Length: 298 amino acids
Theoretical weight: 34.74 KDa
Source organism: Autographa californica nucleopolyhedrovirus
Expression system: Escherichia coli
UniProt:
- Canonical:
 P08160 (Residues: 2-299; Coverage: 100%)
P08160 (Residues: 2-299; Coverage: 100%)
Pfam: Apoptosis preventing protein
InterPro:
CATH: Baculovirus p35
SCOP: Baculovirus p35 protein
Chains: B, D
Length: 258 amino acids
Theoretical weight: 29.52 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
Pfam: Caspase domain
InterPro:
SCOP: Caspase catalytic domain
Length: 258 amino acids
Theoretical weight: 29.52 KDa
Source organism: Homo sapiens
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q14790 (Residues: 222-479; Coverage: 54%)
Q14790 (Residues: 222-479; Coverage: 54%)
Pfam: Caspase domain
InterPro:
- Caspase-like domain superfamily
- Peptidase C14A, caspase catalytic domain
- Peptidase C14, p20 domain
- Peptidase C14, caspase domain
- Peptidase family C14A, His active site
- Peptidase family C14A, cysteine active site
- Peptidase C14, caspase non-catalytic subunit p10
SCOP: Caspase catalytic domain